Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107448 |
| Name | oriT_pRCCGE525b |
| Organism | Rhizobium jaguaris strain CCGE525 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP032696 (528397..528432 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 21..26, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pRCCGE525b
ATCCGAAGGGCGCAATTATACGTCGCTGTCGCGACG
ATCCGAAGGGCGCAATTATACGTCGCTGTCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file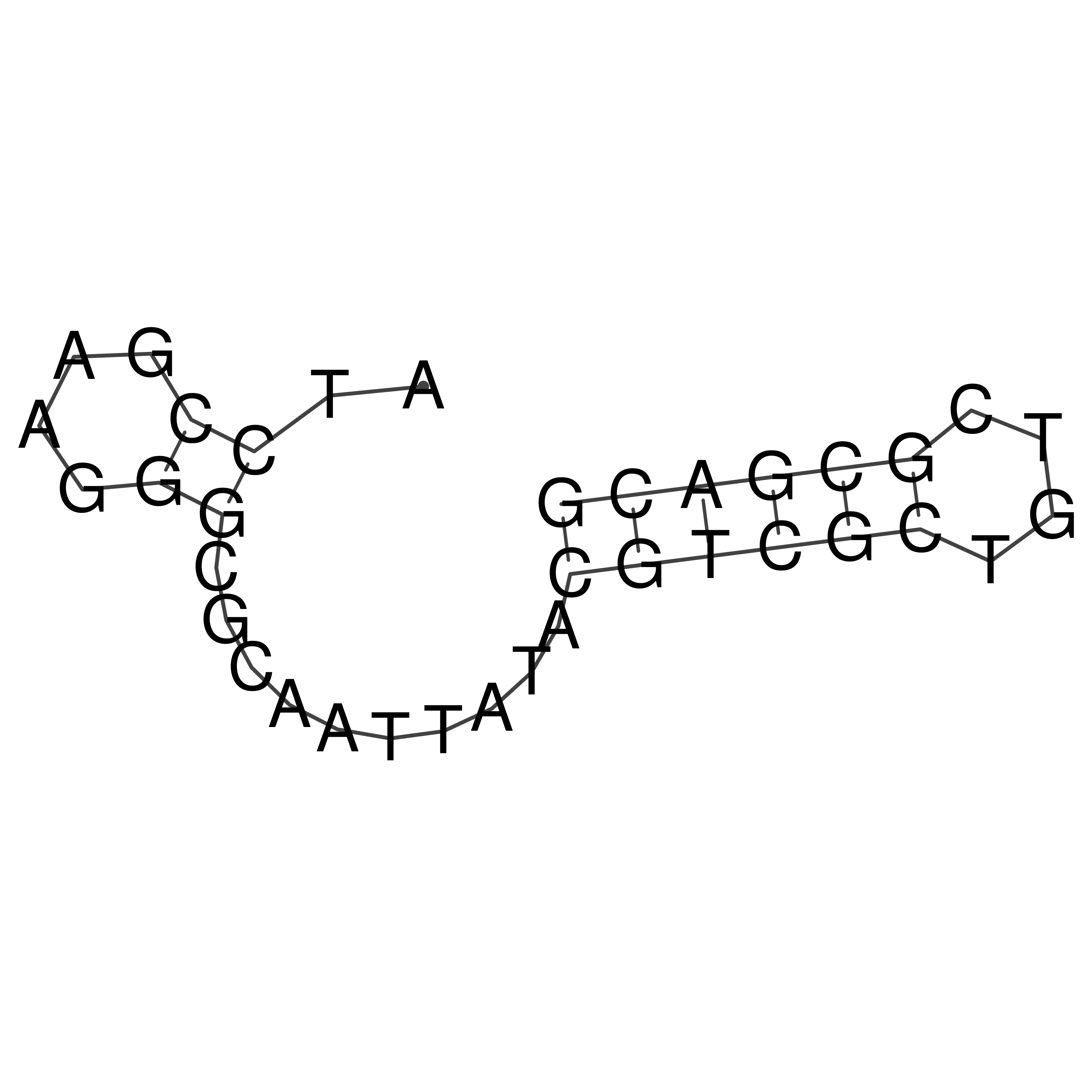
Relaxase
| ID | 4987 | GenBank | WP_120709148 |
| Name | traA_CCGE525_RS36755_pRCCGE525b |
UniProt ID | _ |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122071.79 Da Isoelectric Point: 9.8490
>WP_120709148.1 Ti-type conjugative transfer relaxase TraA [Rhizobium jaguaris]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKEGLLHEEFVIPADAPQWLREMVADR
SVSGASEAFWNSVEVFEKRVDAQLAKDMTIALPMELSTEQNIELVRDFVERHITSAGMVADWVYHDAPGN
PHVHLMTTLRPLAEDGFGAKKVALTGPDGKPIRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIGLTPTIHLGVGAKAIERKSVETDWDISLERLELQDERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQGLMARILQSPDTLRLERERIEFSSGARVPAKFTTRDLIRLEA
EMANRAIWLSQRSSHGVAGPVRQATFARHLHLSDEQKTAIAYATGGARIAAVIGRAGAGKTTTMKAAREA
WETAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRVGYTELETIYRQREQWMRDASLDLARGNIGKAVEAYR
APGHVNGADFKADAVQALIADWNRDYDPAKTSLILAHLRRDVRMLNEMARSKLVERGLVAEGFVFRTEDG
HRKFAAGDQVVFLRNEGSLRVKNGMLAKVVDAAPGRIVAEVGEGEHRRQVTVEQRFYANVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHRDDLGLYYGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
GQALRFAEARGLNLVNVARTVVRDHLEWTVRQKSKLAELGARLAALAGKLGLSAAAVRTSQSHTIKESKP
MVSGITTFPRSLEQAVEDRLAADPGLKKQWEEVSNRFHIVYAQPEAAFKAAKVDTMLKDEAAAKSTIARI
GSEPEGFGALKGKTGILASRADKQNRERAVTNAPALARNLERYLRQRTEAECKYEVEERALRLKVSLDIP
ALTPVAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFARAVSERFGERTFLSLTAKSPNGQTFNA
VTTGMTAGQKAEVGSAWSSMRTVQRLSAHERAAESLTQTETLRQSKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKEGLLHEEFVIPADAPQWLREMVADR
SVSGASEAFWNSVEVFEKRVDAQLAKDMTIALPMELSTEQNIELVRDFVERHITSAGMVADWVYHDAPGN
PHVHLMTTLRPLAEDGFGAKKVALTGPDGKPIRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLALAG
LDIRVDGRSFEKQGIGLTPTIHLGVGAKAIERKSVETDWDISLERLELQDERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQGLMARILQSPDTLRLERERIEFSSGARVPAKFTTRDLIRLEA
EMANRAIWLSQRSSHGVAGPVRQATFARHLHLSDEQKTAIAYATGGARIAAVIGRAGAGKTTTMKAAREA
WETAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRVGYTELETIYRQREQWMRDASLDLARGNIGKAVEAYR
APGHVNGADFKADAVQALIADWNRDYDPAKTSLILAHLRRDVRMLNEMARSKLVERGLVAEGFVFRTEDG
HRKFAAGDQVVFLRNEGSLRVKNGMLAKVVDAAPGRIVAEVGEGEHRRQVTVEQRFYANVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHRDDLGLYYGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
GQALRFAEARGLNLVNVARTVVRDHLEWTVRQKSKLAELGARLAALAGKLGLSAAAVRTSQSHTIKESKP
MVSGITTFPRSLEQAVEDRLAADPGLKKQWEEVSNRFHIVYAQPEAAFKAAKVDTMLKDEAAAKSTIARI
GSEPEGFGALKGKTGILASRADKQNRERAVTNAPALARNLERYLRQRTEAECKYEVEERALRLKVSLDIP
ALTPVAKQTLERVRDAIDRNDLPAGLEYALADKMVKAELEGFARAVSERFGERTFLSLTAKSPNGQTFNA
VTTGMTAGQKAEVGSAWSSMRTVQRLSAHERAAESLTQTETLRQSKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5195 | GenBank | WP_120709145 |
| Name | traG_CCGE525_RS36740_pRCCGE525b |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 69775.91 Da Isoelectric Point: 9.1987
>WP_120709145.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTRILLFVVPCALMISTAIGMTGIEKWLSAFGKTDTARQILGRVGIALPYIVASFEGIVLLFASAGSVRI
RTAGWGVVAGAIATVLVAVLREAMRLSAFREQVPAGRFLFNYLDPATAIGAAVVLMSALFGMRVVIAGNA
AFAKAEPKRIYGKRALHGEADWMKLSQAEQLFPEDGGIVIGERYRVDTDSVAAQSFRADRAETWGAGGTS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALIILDPSNEVAPMASKHRGGNRDVFILDPKH
SEIGFNALDWIGRFGGTKEEDIASVASWVVSDSGARGLRDDFFRASALQLLTALIADVCLSGHTIENDQT
LRQVRKNLSEPEPKLRERLQSIYDNSESDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAALVS
GTTFTTGDLAGGKTDVFINIDLKTLEMHSGLARVIIGSFLNAMCNLNGEIQGRTLFLLDEVARLGYMRIL
ETARDAGRKYGITLTMIYQSIGQMRETYGGRDATSKWFESASWISFAAINDPETADYISRRSGMTTVEID
QVSRSFQSRGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNPALRCGRAIWFRRRDMKDCINGKSF
QWGRKSITL
MTRILLFVVPCALMISTAIGMTGIEKWLSAFGKTDTARQILGRVGIALPYIVASFEGIVLLFASAGSVRI
RTAGWGVVAGAIATVLVAVLREAMRLSAFREQVPAGRFLFNYLDPATAIGAAVVLMSALFGMRVVIAGNA
AFAKAEPKRIYGKRALHGEADWMKLSQAEQLFPEDGGIVIGERYRVDTDSVAAQSFRADRAETWGAGGTS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALIILDPSNEVAPMASKHRGGNRDVFILDPKH
SEIGFNALDWIGRFGGTKEEDIASVASWVVSDSGARGLRDDFFRASALQLLTALIADVCLSGHTIENDQT
LRQVRKNLSEPEPKLRERLQSIYDNSESDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAALVS
GTTFTTGDLAGGKTDVFINIDLKTLEMHSGLARVIIGSFLNAMCNLNGEIQGRTLFLLDEVARLGYMRIL
ETARDAGRKYGITLTMIYQSIGQMRETYGGRDATSKWFESASWISFAAINDPETADYISRRSGMTTVEID
QVSRSFQSRGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNPALRCGRAIWFRRRDMKDCINGKSF
QWGRKSITL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5196 | GenBank | WP_120709159 |
| Name | t4cp2_CCGE525_RS36825_pRCCGE525b |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91980.91 Da Isoelectric Point: 5.9945
>WP_120709159.1 conjugal transfer protein TrbE [Rhizobium jaguaris]
MVALKHFRTTGPSFADLVPYAGLIDNGVLLLKDGSLMAGWYFAGPDCESATDFERNELSRQINTVLSRLG
SGWMIQVEAVRIPTFDYPSDERCHFPDAVTRAIDVERRAHFAREHGHFESKHALVLTYRPLESKKTALSK
YIYSDEESRKKTYADTVLFIFKNAVREIEQYLANTLSIRRMGTRETVERGGERVARYDELLQFVRFCITG
ENHPIRLPEAPMYLDWIATAELEHGLTPKVESRFLGVIAIDGLPAESCPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQRGSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVIVL
FDRDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLVPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYDQAQIFA
FDKGNSLLPLTLATGGDHYEIGGDVQEEGRALAFCPLADLSGDADRAWAVEWIEMLVALQSVTITPDHRN
AISRQISLMASARGRSLSDFVSGVQMREIKDALYQYTVDGPMGQLLDAEEDGLSLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNQRQIEIVSNAIPKREYYVVTPEGRRLFDMAPGP
VALSFIGASGKHDLKRIRTLSSEHGRDWPIHWLQMRGVHDAASLLNFE
MVALKHFRTTGPSFADLVPYAGLIDNGVLLLKDGSLMAGWYFAGPDCESATDFERNELSRQINTVLSRLG
SGWMIQVEAVRIPTFDYPSDERCHFPDAVTRAIDVERRAHFAREHGHFESKHALVLTYRPLESKKTALSK
YIYSDEESRKKTYADTVLFIFKNAVREIEQYLANTLSIRRMGTRETVERGGERVARYDELLQFVRFCITG
ENHPIRLPEAPMYLDWIATAELEHGLTPKVESRFLGVIAIDGLPAESCPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQRGSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVIVL
FDRDREALQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLVPLNSVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYDQAQIFA
FDKGNSLLPLTLATGGDHYEIGGDVQEEGRALAFCPLADLSGDADRAWAVEWIEMLVALQSVTITPDHRN
AISRQISLMASARGRSLSDFVSGVQMREIKDALYQYTVDGPMGQLLDAEEDGLSLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNQRQIEIVSNAIPKREYYVVTPEGRRLFDMAPGP
VALSFIGASGKHDLKRIRTLSSEHGRDWPIHWLQMRGVHDAASLLNFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 533554..545882
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CCGE525_RS36760 (CCGE525_36735) | 531896..532384 | + | 489 | WP_245472478 | conjugative transfer signal peptidase TraF | - |
| CCGE525_RS36765 (CCGE525_36740) | 532374..533537 | + | 1164 | WP_120709150 | conjugal transfer protein TraB | - |
| CCGE525_RS36770 (CCGE525_36745) | 533554..534162 | + | 609 | WP_120709151 | TraH family protein | virB1 |
| CCGE525_RS36780 (CCGE525_36755) | 534873..535109 | - | 237 | WP_120709152 | helix-turn-helix transcriptional regulator | - |
| CCGE525_RS36785 (CCGE525_36760) | 535359..535673 | + | 315 | WP_120709153 | transcriptional repressor TraM | - |
| CCGE525_RS36790 (CCGE525_36765) | 535677..536381 | - | 705 | WP_120709220 | autoinducer binding domain-containing protein | - |
| CCGE525_RS36795 (CCGE525_36770) | 536539..537840 | - | 1302 | WP_120709154 | IncP-type conjugal transfer protein TrbI | virB10 |
| CCGE525_RS36800 (CCGE525_36775) | 537853..538290 | - | 438 | WP_120709155 | conjugal transfer protein TrbH | - |
| CCGE525_RS36805 (CCGE525_36780) | 538294..539112 | - | 819 | WP_120709156 | P-type conjugative transfer protein TrbG | virB9 |
| CCGE525_RS36810 (CCGE525_36785) | 539130..539792 | - | 663 | WP_120709157 | conjugal transfer protein TrbF | virB8 |
| CCGE525_RS36815 (CCGE525_36790) | 539817..540995 | - | 1179 | WP_120709158 | P-type conjugative transfer protein TrbL | virB6 |
| CCGE525_RS36820 (CCGE525_36795) | 541000..541802 | - | 803 | Protein_507 | P-type conjugative transfer protein TrbJ | - |
| CCGE525_RS36825 (CCGE525_36800) | 541774..544230 | - | 2457 | WP_120709159 | conjugal transfer protein TrbE | virb4 |
| CCGE525_RS36830 (CCGE525_36805) | 544240..544539 | - | 300 | WP_120709160 | conjugal transfer protein TrbD | virB3 |
| CCGE525_RS36835 (CCGE525_36810) | 544532..544915 | - | 384 | WP_120709161 | TrbC/VirB2 family protein | virB2 |
| CCGE525_RS36840 (CCGE525_36815) | 544905..545882 | - | 978 | WP_120709162 | P-type conjugative transfer ATPase TrbB | virB11 |
| CCGE525_RS36845 (CCGE525_36820) | 545879..546517 | - | 639 | WP_120709163 | acyl-homoserine-lactone synthase TraI | - |
| CCGE525_RS36850 (CCGE525_36825) | 546893..548113 | + | 1221 | WP_120709164 | plasmid partitioning protein RepA | - |
| CCGE525_RS36855 (CCGE525_36830) | 548173..549147 | + | 975 | WP_120709165 | plasmid partitioning protein RepB | - |
| CCGE525_RS36860 (CCGE525_36835) | 549307..550563 | + | 1257 | WP_120709166 | plasmid replication protein RepC | - |
Host bacterium
| ID | 7883 | GenBank | NZ_CP032696 |
| Plasmid name | pRCCGE525b | Incompatibility group | - |
| Plasmid size | 550563 bp | Coordinate of oriT [Strand] | 528397..528432 [+] |
| Host baterium | Rhizobium jaguaris strain CCGE525 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | nodD, nodA, nodE, fixN, fixO, nodB, nodC, nodS, nodU, nodI, nodJ, nodH, nifT, nifZ, nifB, nifA, fixX, fixC, fixB, fixA, nifW, nifS, nifH, nifD, nifE, nifN, nifX, mLTONO_5203, nodM |
| Anti-CRISPR | - |