Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107441 |
| Name | oriT_pRCCGE531b |
| Organism | Rhizobium sp. CCGE531 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP032687 (555923..555958 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 21..26, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pRCCGE531b
ATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
ATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file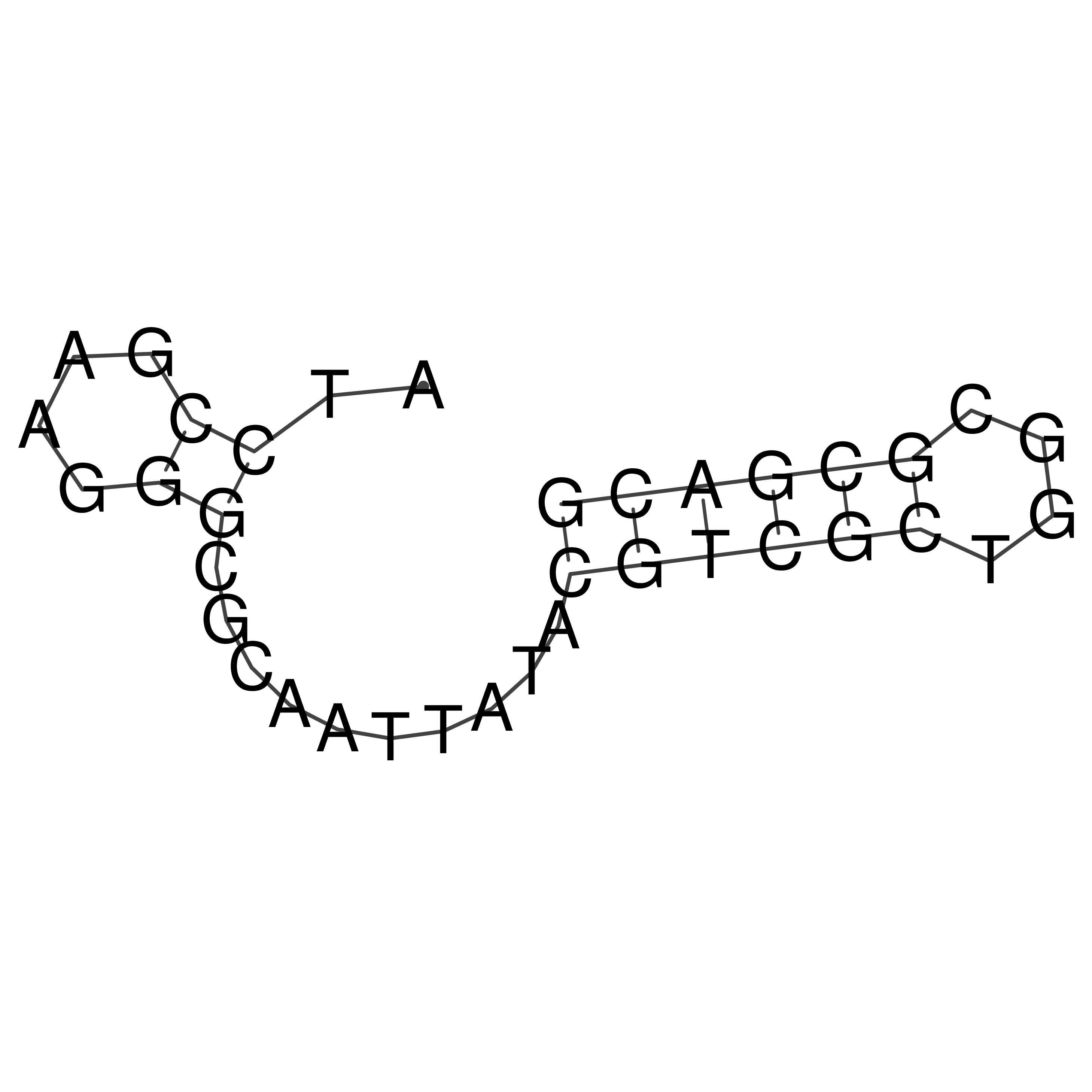
Relaxase
| ID | 4977 | GenBank | WP_065091806 |
| Name | traA_CCGE531_RS32575_pRCCGE531b |
UniProt ID | A0A387HA03 |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122065.88 Da Isoelectric Point: 10.0272
>WP_065091806.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADAPSWLRTMIADR
SVSGASEAFWNSVEIFEKRVDAQLAKDVTVALPMELSSEQNIELVRDFIERHITSKGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPGGKPVRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLARAG
LDLRVDGHSFEKQGINLTPTIHLGVGAKAIERKSAEAERAPSLERLELQEERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQSLMARILQSPETLRLERERIEFSGGLRVPTKYTTRDLVRLEA
EMANRAIWLAQRSSHGVADAVREATFARHLRLADEQKTAIAHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYGELENIYRQREQWMREASLDLARGNIGRAVEAYR
AQGRVSGADLKADAVQALIADWNRDYDPQRSSLILAHLRRDVRMLNGMARAKLVERGLVADGFAFRTEDG
HRKFAAGDQIVFLKNEGSLGVKNGMLAKVVDATPGRIVAEVGDGEHRRQVTVEQRFYANLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDFGLYHGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
AQALRFADARGLHLVNVARTVVRDRIEWTVRQKSKLAALGARLAAIARRLGLSAGAAKTSQSHSMQEAKP
MVSGITTFPKSLEQVVEDLLAADPGLKKQWEEVSARFHLVYAQPEDAFKAVNVDAMLRNEAAAKSTITKI
GSEPETFGALKGKTGILAGRADKQAHERAVTNAPALARNLERYLRQRTEAEHKYDAEERAVRMKISLDIP
ALSPAAKQTLERVRDAIDRNDLPAGLENALADKMVKAELEGFARAVSERFGERTFLPLAAKDMTGQTFQS
VTSSMSAGQKAEVQSAWSTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADAPSWLRTMIADR
SVSGASEAFWNSVEIFEKRVDAQLAKDVTVALPMELSSEQNIELVRDFIERHITSKGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPGGKPVRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLARAG
LDLRVDGHSFEKQGINLTPTIHLGVGAKAIERKSAEAERAPSLERLELQEERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQSLMARILQSPETLRLERERIEFSGGLRVPTKYTTRDLVRLEA
EMANRAIWLAQRSSHGVADAVREATFARHLRLADEQKTAIAHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYGELENIYRQREQWMREASLDLARGNIGRAVEAYR
AQGRVSGADLKADAVQALIADWNRDYDPQRSSLILAHLRRDVRMLNGMARAKLVERGLVADGFAFRTEDG
HRKFAAGDQIVFLKNEGSLGVKNGMLAKVVDATPGRIVAEVGDGEHRRQVTVEQRFYANLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDFGLYHGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
AQALRFADARGLHLVNVARTVVRDRIEWTVRQKSKLAALGARLAAIARRLGLSAGAAKTSQSHSMQEAKP
MVSGITTFPKSLEQVVEDLLAADPGLKKQWEEVSARFHLVYAQPEDAFKAVNVDAMLRNEAAAKSTITKI
GSEPETFGALKGKTGILAGRADKQAHERAVTNAPALARNLERYLRQRTEAEHKYDAEERAVRMKISLDIP
ALSPAAKQTLERVRDAIDRNDLPAGLENALADKMVKAELEGFARAVSERFGERTFLPLAAKDMTGQTFQS
VTSSMSAGQKAEVQSAWSTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A387HA03 |
T4CP
| ID | 5179 | GenBank | WP_004119814 |
| Name | traG_CCGE531_RS32560_pRCCGE531b |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 71115.08 Da Isoelectric Point: 9.0145
>WP_004119814.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTRILLFVAPCALMMLMAIGMTGTELWLSHFGKSDAARQMLGRAGIALPYVAASVVGIIFLFASAGSARI
RTAGWGVVTGATATLVLAVLREASRLSAYLEQVPAGKTVFNYLDPATAIGAAAVLMSALFGMRVAIAGNA
TFARAEPKRIVGKRALHGEADWMKLSQAEKLFAESGGIVIGERYRVDRDSIAAPSFRADSAASWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVLKHRGDANRDVFVLDPK
HSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTTEND
QTLRQVRKNLSEPEPQLRQRLHEIYDDSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAAL
VSGTSFSTSDLADGKTDVFINIDLKTLETHAGLARVIIGAFLNAIYNRNGEMQGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKGCVGAR
AFSKAANPAENSPDRPARSAASKADREQ
MTRILLFVAPCALMMLMAIGMTGTELWLSHFGKSDAARQMLGRAGIALPYVAASVVGIIFLFASAGSARI
RTAGWGVVTGATATLVLAVLREASRLSAYLEQVPAGKTVFNYLDPATAIGAAAVLMSALFGMRVAIAGNA
TFARAEPKRIVGKRALHGEADWMKLSQAEKLFAESGGIVIGERYRVDRDSIAAPSFRADSAASWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVLKHRGDANRDVFVLDPK
HSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTTEND
QTLRQVRKNLSEPEPQLRQRLHEIYDDSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAAL
VSGTSFSTSDLADGKTDVFINIDLKTLETHAGLARVIIGAFLNAIYNRNGEMQGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKGCVGAR
AFSKAANPAENSPDRPARSAASKADREQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5180 | GenBank | WP_065091804 |
| Name | t4cp2_CCGE531_RS32640_pRCCGE531b |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91755.65 Da Isoelectric Point: 6.3779
>WP_065091804.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRAPDPSFADLVPYAGLVDNGVLLLKDGSLMAAWYFAGPDSESATALDRNELSRHINTILSRLG
SGWMIQVEAVRIPTFDYPSDNRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEVSRKKTYADTVLFIFKNAVREIEQYFANTLSIRRMETREALERGGARVARYDELLQFVRFCITG
DNHPIRLPDAPMYLDWIATAELEHGLTSKVESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMNMVAETEEAIAQASSQLVAYGYYTPVIVL
FDHDREGLQEKAESIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYERAQIFA
FDKGNSLLPLTLATGGDHYEIGGDAQEEAKALAFCPLSDLSSDSDRAWAAEWIEMLIALQGVTITPDHRN
AVSRQIGLMASAAGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEKDGLLLGSFQTFEIEELMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAASEPGTREFYERIGFNERQIEIVSNAIPKREYYVATSEGRRLFDMSLGP
VALSFVGASGKEDLKRIRTLSSKHGRDWPIHWLRTRGVQDAAALLNLE
MVALKRFRAPDPSFADLVPYAGLVDNGVLLLKDGSLMAAWYFAGPDSESATALDRNELSRHINTILSRLG
SGWMIQVEAVRIPTFDYPSDNRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEVSRKKTYADTVLFIFKNAVREIEQYFANTLSIRRMETREALERGGARVARYDELLQFVRFCITG
DNHPIRLPDAPMYLDWIATAELEHGLTSKVESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMNMVAETEEAIAQASSQLVAYGYYTPVIVL
FDHDREGLQEKAESIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYERAQIFA
FDKGNSLLPLTLATGGDHYEIGGDAQEEAKALAFCPLSDLSSDSDRAWAAEWIEMLIALQGVTITPDHRN
AVSRQIGLMASAAGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEKDGLLLGSFQTFEIEELMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAASEPGTREFYERIGFNERQIEIVSNAIPKREYYVATSEGRRLFDMSLGP
VALSFVGASGKEDLKRIRTLSSKHGRDWPIHWLRTRGVQDAAALLNLE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 561081..572952
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CCGE531_RS32580 (CCGE531_32595) | 559346..559912 | + | 567 | WP_120670997 | conjugative transfer signal peptidase TraF | - |
| CCGE531_RS32585 (CCGE531_32600) | 559902..561065 | + | 1164 | WP_004119833 | conjugal transfer protein TraB | - |
| CCGE531_RS32590 (CCGE531_32605) | 561081..561689 | + | 609 | WP_004119835 | TraH family protein | virB1 |
| CCGE531_RS32595 (CCGE531_32610) | 561953..562189 | - | 237 | WP_004119837 | helix-turn-helix transcriptional regulator | - |
| CCGE531_RS32600 (CCGE531_32615) | 562439..562753 | + | 315 | WP_065091805 | transcriptional repressor TraM | - |
| CCGE531_RS32605 (CCGE531_32620) | 562757..563461 | - | 705 | WP_004119842 | autoinducer binding domain-containing protein | - |
| CCGE531_RS32610 (CCGE531_32625) | 563620..564921 | - | 1302 | WP_004119844 | IncP-type conjugal transfer protein TrbI | virB10 |
| CCGE531_RS32615 (CCGE531_32630) | 564932..565366 | - | 435 | WP_004119847 | conjugal transfer protein TrbH | - |
| CCGE531_RS32620 (CCGE531_32635) | 565370..566182 | - | 813 | WP_004119849 | P-type conjugative transfer protein TrbG | virB9 |
| CCGE531_RS32625 (CCGE531_32640) | 566199..566861 | - | 663 | WP_004119850 | conjugal transfer protein TrbF | virB8 |
| CCGE531_RS32630 (CCGE531_32645) | 566886..568064 | - | 1179 | WP_004119851 | P-type conjugative transfer protein TrbL | virB6 |
| CCGE531_RS32635 (CCGE531_32650) | 568069..568872 | - | 804 | WP_004119852 | P-type conjugative transfer protein TrbJ | virB5 |
| CCGE531_RS32640 (CCGE531_32655) | 568844..571300 | - | 2457 | WP_065091804 | conjugal transfer protein TrbE | virb4 |
| CCGE531_RS32645 (CCGE531_32660) | 571310..571609 | - | 300 | WP_004119854 | conjugal transfer protein TrbD | virB3 |
| CCGE531_RS32650 (CCGE531_32665) | 571602..571985 | - | 384 | WP_004119857 | TrbC/VirB2 family protein | virB2 |
| CCGE531_RS32655 (CCGE531_32670) | 571975..572952 | - | 978 | WP_004119858 | P-type conjugative transfer ATPase TrbB | virB11 |
| CCGE531_RS32660 (CCGE531_32675) | 572949..573587 | - | 639 | WP_004119859 | acyl-homoserine-lactone synthase TraI | - |
| CCGE531_RS32665 (CCGE531_32680) | 573966..575186 | + | 1221 | WP_028755142 | plasmid partitioning protein RepA | - |
| CCGE531_RS32670 (CCGE531_32685) | 575246..576220 | + | 975 | WP_004119861 | plasmid partitioning protein RepB | - |
| CCGE531_RS32675 (CCGE531_32690) | 576382..577596 | + | 1215 | WP_120670998 | plasmid replication protein RepC | - |
Host bacterium
| ID | 7876 | GenBank | NZ_CP032687 |
| Plasmid name | pRCCGE531b | Incompatibility group | - |
| Plasmid size | 577596 bp | Coordinate of oriT [Strand] | 555923..555958 [+] |
| Host baterium | Rhizobium sp. CCGE531 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | LinG, dxnH, linJ |
| Symbiosis gene | nodD, nodA, nodE, fixN, fixO, fixQ, fixG, nodB, nodC, nodS, nodU, nodI, nodJ, nodH, nifT, nifZ, nifB, nifA, fixX, fixC, fixB, fixA, nifW, nifS, nifH, nifD, nifK, nifE, nifN, nifX, mLTONO_5203, nodM |
| Anti-CRISPR | AcrIIA7 |