Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107440 |
| Name | oriT_pRCCGE532b |
| Organism | Rhizobium sp. CCGE532 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP032692 (484487..484522 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 21..26, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pRCCGE532b
ATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
ATCCGAAGGGCGCAATTATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file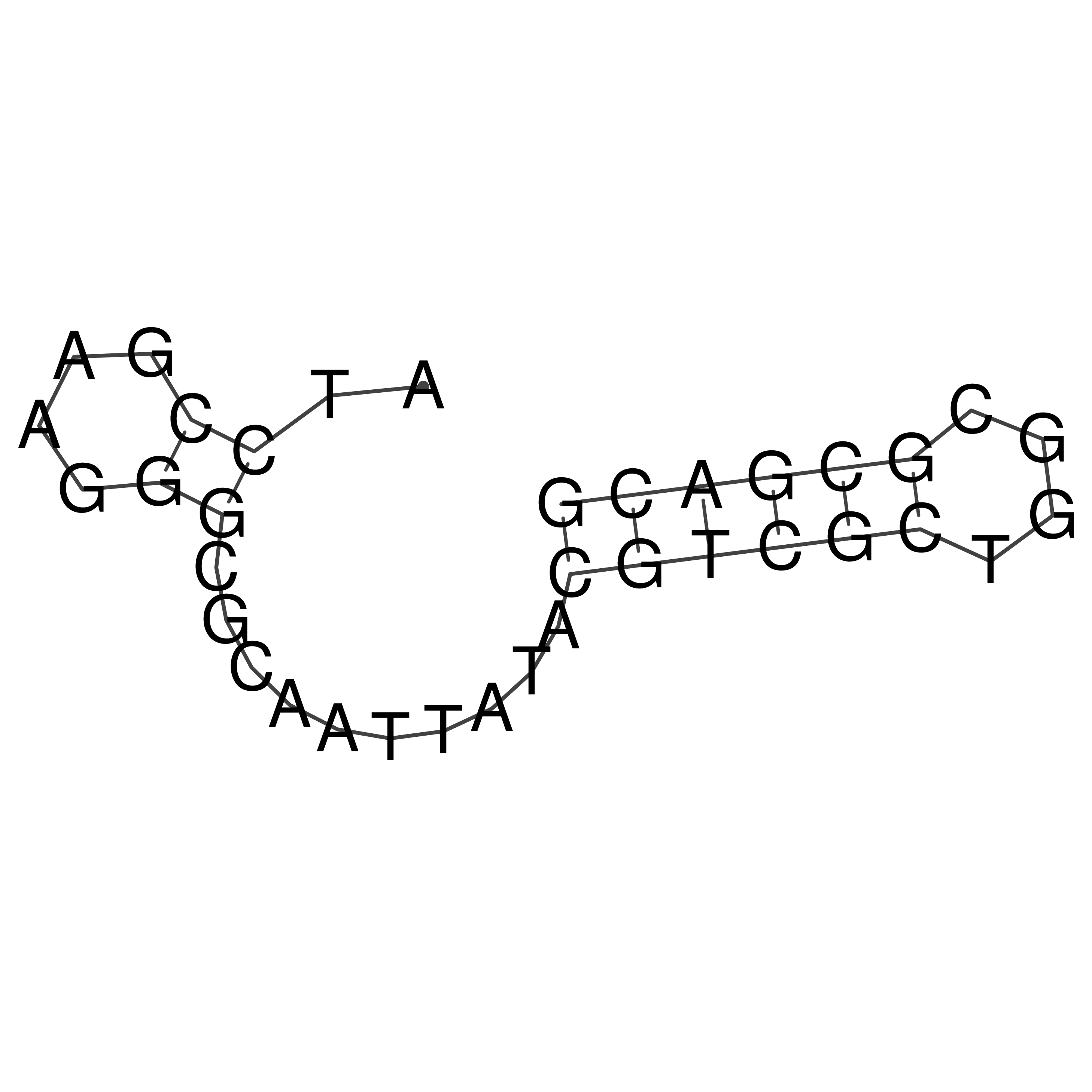
Relaxase
| ID | 4976 | GenBank | WP_065091806 |
| Name | traA_CCGE532_RS31740_pRCCGE532b |
UniProt ID | A0A387HA03 |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122065.88 Da Isoelectric Point: 10.0272
>WP_065091806.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADAPSWLRTMIADR
SVSGASEAFWNSVEIFEKRVDAQLAKDVTVALPMELSSEQNIELVRDFIERHITSKGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPGGKPVRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLARAG
LDLRVDGHSFEKQGINLTPTIHLGVGAKAIERKSAEAERAPSLERLELQEERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQSLMARILQSPETLRLERERIEFSGGLRVPTKYTTRDLVRLEA
EMANRAIWLAQRSSHGVADAVREATFARHLRLADEQKTAIAHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYGELENIYRQREQWMREASLDLARGNIGRAVEAYR
AQGRVSGADLKADAVQALIADWNRDYDPQRSSLILAHLRRDVRMLNGMARAKLVERGLVADGFAFRTEDG
HRKFAAGDQIVFLKNEGSLGVKNGMLAKVVDATPGRIVAEVGDGEHRRQVTVEQRFYANLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDFGLYHGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
AQALRFADARGLHLVNVARTVVRDRIEWTVRQKSKLAALGARLAAIARRLGLSAGAAKTSQSHSMQEAKP
MVSGITTFPKSLEQVVEDLLAADPGLKKQWEEVSARFHLVYAQPEDAFKAVNVDAMLRNEAAAKSTITKI
GSEPETFGALKGKTGILAGRADKQAHERAVTNAPALARNLERYLRQRTEAEHKYDAEERAVRMKISLDIP
ALSPAAKQTLERVRDAIDRNDLPAGLENALADKMVKAELEGFARAVSERFGERTFLPLAAKDMTGQTFQS
VTSSMSAGQKAEVQSAWSTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVIPADAPSWLRTMIADR
SVSGASEAFWNSVEIFEKRVDAQLAKDVTVALPMELSSEQNIELVRDFIERHITSKGMIADWVYHDAPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPGGKPVRNDAGKIVYELWAGSLDDFNAFRDGWFACQNRHLARAG
LDLRVDGHSFEKQGINLTPTIHLGVGAKAIERKSAEAERAPSLERLELQEERRAENARRIQRRPELVLDL
ITREKSVFDERDVAKILHRYIDDAGLFQSLMARILQSPETLRLERERIEFSGGLRVPTKYTTRDLVRLEA
EMANRAIWLAQRSSHGVADAVREATFARHLRLADEQKTAIAHVTGGERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAEGLEKEAGILSRTLSSWELRWNQGRNQLDDKTVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYGELENIYRQREQWMREASLDLARGNIGRAVEAYR
AQGRVSGADLKADAVQALIADWNRDYDPQRSSLILAHLRRDVRMLNGMARAKLVERGLVADGFAFRTEDG
HRKFAAGDQIVFLKNEGSLGVKNGMLAKVVDATPGRIVAEVGDGEHRRQVTVEQRFYANLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDFGLYHGRRSFAKAGGLIPILSRRQAKETTLDYASGRFY
AQALRFADARGLHLVNVARTVVRDRIEWTVRQKSKLAALGARLAAIARRLGLSAGAAKTSQSHSMQEAKP
MVSGITTFPKSLEQVVEDLLAADPGLKKQWEEVSARFHLVYAQPEDAFKAVNVDAMLRNEAAAKSTITKI
GSEPETFGALKGKTGILAGRADKQAHERAVTNAPALARNLERYLRQRTEAEHKYDAEERAVRMKISLDIP
ALSPAAKQTLERVRDAIDRNDLPAGLENALADKMVKAELEGFARAVSERFGERTFLPLAAKDMTGQTFQS
VTSSMSAGQKAEVQSAWSTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A387HA03 |
T4CP
| ID | 5177 | GenBank | WP_004119814 |
| Name | traG_CCGE532_RS31725_pRCCGE532b |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 71115.08 Da Isoelectric Point: 9.0145
>WP_004119814.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MTRILLFVAPCALMMLMAIGMTGTELWLSHFGKSDAARQMLGRAGIALPYVAASVVGIIFLFASAGSARI
RTAGWGVVTGATATLVLAVLREASRLSAYLEQVPAGKTVFNYLDPATAIGAAAVLMSALFGMRVAIAGNA
TFARAEPKRIVGKRALHGEADWMKLSQAEKLFAESGGIVIGERYRVDRDSIAAPSFRADSAASWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVLKHRGDANRDVFVLDPK
HSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTTEND
QTLRQVRKNLSEPEPQLRQRLHEIYDDSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAAL
VSGTSFSTSDLADGKTDVFINIDLKTLETHAGLARVIIGAFLNAIYNRNGEMQGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKGCVGAR
AFSKAANPAENSPDRPARSAASKADREQ
MTRILLFVAPCALMMLMAIGMTGTELWLSHFGKSDAARQMLGRAGIALPYVAASVVGIIFLFASAGSARI
RTAGWGVVTGATATLVLAVLREASRLSAYLEQVPAGKTVFNYLDPATAIGAAAVLMSALFGMRVAIAGNA
TFARAEPKRIVGKRALHGEADWMKLSQAEKLFAESGGIVIGERYRVDRDSIAAPSFRADSAASWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGALVVLDPSNEVAPMVLKHRGDANRDVFVLDPK
HSEIGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTTEND
QTLRQVRKNLSEPEPQLRQRLHEIYDDSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYAAL
VSGTSFSTSDLADGKTDVFINIDLKTLETHAGLARVIIGAFLNAIYNRNGEMQGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSVQSKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKGCVGAR
AFSKAANPAENSPDRPARSAASKADREQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5178 | GenBank | WP_065091804 |
| Name | t4cp2_CCGE532_RS31805_pRCCGE532b |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91755.65 Da Isoelectric Point: 6.3779
>WP_065091804.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium]
MVALKRFRAPDPSFADLVPYAGLVDNGVLLLKDGSLMAAWYFAGPDSESATALDRNELSRHINTILSRLG
SGWMIQVEAVRIPTFDYPSDNRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEVSRKKTYADTVLFIFKNAVREIEQYFANTLSIRRMETREALERGGARVARYDELLQFVRFCITG
DNHPIRLPDAPMYLDWIATAELEHGLTSKVESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMNMVAETEEAIAQASSQLVAYGYYTPVIVL
FDHDREGLQEKAESIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYERAQIFA
FDKGNSLLPLTLATGGDHYEIGGDAQEEAKALAFCPLSDLSSDSDRAWAAEWIEMLIALQGVTITPDHRN
AVSRQIGLMASAAGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEKDGLLLGSFQTFEIEELMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAASEPGTREFYERIGFNERQIEIVSNAIPKREYYVATSEGRRLFDMSLGP
VALSFVGASGKEDLKRIRTLSSKHGRDWPIHWLRTRGVQDAAALLNLE
MVALKRFRAPDPSFADLVPYAGLVDNGVLLLKDGSLMAAWYFAGPDSESATALDRNELSRHINTILSRLG
SGWMIQVEAVRIPTFDYPSDNRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDEVSRKKTYADTVLFIFKNAVREIEQYFANTLSIRRMETREALERGGARVARYDELLQFVRFCITG
DNHPIRLPDAPMYLDWIATAELEHGLTSKVESRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMNMVAETEEAIAQASSQLVAYGYYTPVIVL
FDHDREGLQEKAESIRRLIQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTSNLADLIPLNSVWS
GSPVAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYERAQIFA
FDKGNSLLPLTLATGGDHYEIGGDAQEEAKALAFCPLSDLSSDSDRAWAAEWIEMLIALQGVTITPDHRN
AVSRQIGLMASAAGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEKDGLLLGSFQTFEIEELMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAASEPGTREFYERIGFNERQIEIVSNAIPKREYYVATSEGRRLFDMSLGP
VALSFVGASGKEDLKRIRTLSSKHGRDWPIHWLRTRGVQDAAALLNLE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 489645..501516
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CCGE532_RS31745 (CCGE532_31750) | 487910..488476 | + | 567 | WP_004119831 | conjugative transfer signal peptidase TraF | - |
| CCGE532_RS31750 (CCGE532_31755) | 488466..489629 | + | 1164 | WP_004119833 | conjugal transfer protein TraB | - |
| CCGE532_RS31755 (CCGE532_31760) | 489645..490253 | + | 609 | WP_004119835 | TraH family protein | virB1 |
| CCGE532_RS31760 (CCGE532_31765) | 490517..490753 | - | 237 | WP_004119837 | helix-turn-helix transcriptional regulator | - |
| CCGE532_RS31765 (CCGE532_31770) | 491003..491317 | + | 315 | WP_004119838 | transcriptional repressor TraM | - |
| CCGE532_RS31770 (CCGE532_31775) | 491321..492025 | - | 705 | WP_004119842 | autoinducer binding domain-containing protein | - |
| CCGE532_RS31775 (CCGE532_31780) | 492184..493485 | - | 1302 | WP_004119844 | IncP-type conjugal transfer protein TrbI | virB10 |
| CCGE532_RS31780 (CCGE532_31785) | 493496..493930 | - | 435 | WP_004119847 | conjugal transfer protein TrbH | - |
| CCGE532_RS31785 (CCGE532_31790) | 493934..494746 | - | 813 | WP_004119849 | P-type conjugative transfer protein TrbG | virB9 |
| CCGE532_RS31790 (CCGE532_31795) | 494763..495425 | - | 663 | WP_004119850 | conjugal transfer protein TrbF | virB8 |
| CCGE532_RS31795 (CCGE532_31800) | 495450..496628 | - | 1179 | WP_004119851 | P-type conjugative transfer protein TrbL | virB6 |
| CCGE532_RS31800 (CCGE532_31805) | 496633..497436 | - | 804 | WP_004119852 | P-type conjugative transfer protein TrbJ | virB5 |
| CCGE532_RS31805 (CCGE532_31810) | 497408..499864 | - | 2457 | WP_065091804 | conjugal transfer protein TrbE | virb4 |
| CCGE532_RS31810 (CCGE532_31815) | 499874..500173 | - | 300 | WP_004119854 | conjugal transfer protein TrbD | virB3 |
| CCGE532_RS31815 (CCGE532_31820) | 500166..500549 | - | 384 | WP_004119857 | TrbC/VirB2 family protein | virB2 |
| CCGE532_RS31820 (CCGE532_31825) | 500539..501516 | - | 978 | WP_004119858 | P-type conjugative transfer ATPase TrbB | virB11 |
| CCGE532_RS31825 (CCGE532_31830) | 501513..502151 | - | 639 | WP_004119859 | acyl-homoserine-lactone synthase TraI | - |
| CCGE532_RS31830 (CCGE532_31835) | 502530..503750 | + | 1221 | WP_028755142 | plasmid partitioning protein RepA | - |
| CCGE532_RS31835 (CCGE532_31840) | 503810..504784 | + | 975 | WP_004119861 | plasmid partitioning protein RepB | - |
| CCGE532_RS31840 (CCGE532_31845) | 504946..506160 | + | 1215 | WP_120764334 | plasmid replication protein RepC | - |
Host bacterium
| ID | 7875 | GenBank | NZ_CP032692 |
| Plasmid name | pRCCGE532b | Incompatibility group | - |
| Plasmid size | 506160 bp | Coordinate of oriT [Strand] | 484487..484522 [+] |
| Host baterium | Rhizobium sp. CCGE532 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | linJ, dxnH, LinG |
| Symbiosis gene | nodD, nodA, mLTONO_5203, nifX, nifN, nifE, nifK, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifB, nifZ, nifT, nodH, nodJ, nodI, nodU, nodS, nodC, nodB, fixG, fixQ, fixO, fixN, nodE, nodM |
| Anti-CRISPR | - |