Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107349 |
| Name | oriT_ISRA436|unnamed |
| Organism | Bradyrhizobium sp. ISRA436 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP094520 (84507..84544 [-], 38 nt) |
| oriT length | 38 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 38 nt
>oriT_ISRA436|unnamed
TCCAAGGGCGCAGTAATACGTCGCTGACGCGACGTCCT
TCCAAGGGCGCAGTAATACGTCGCTGACGCGACGTCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file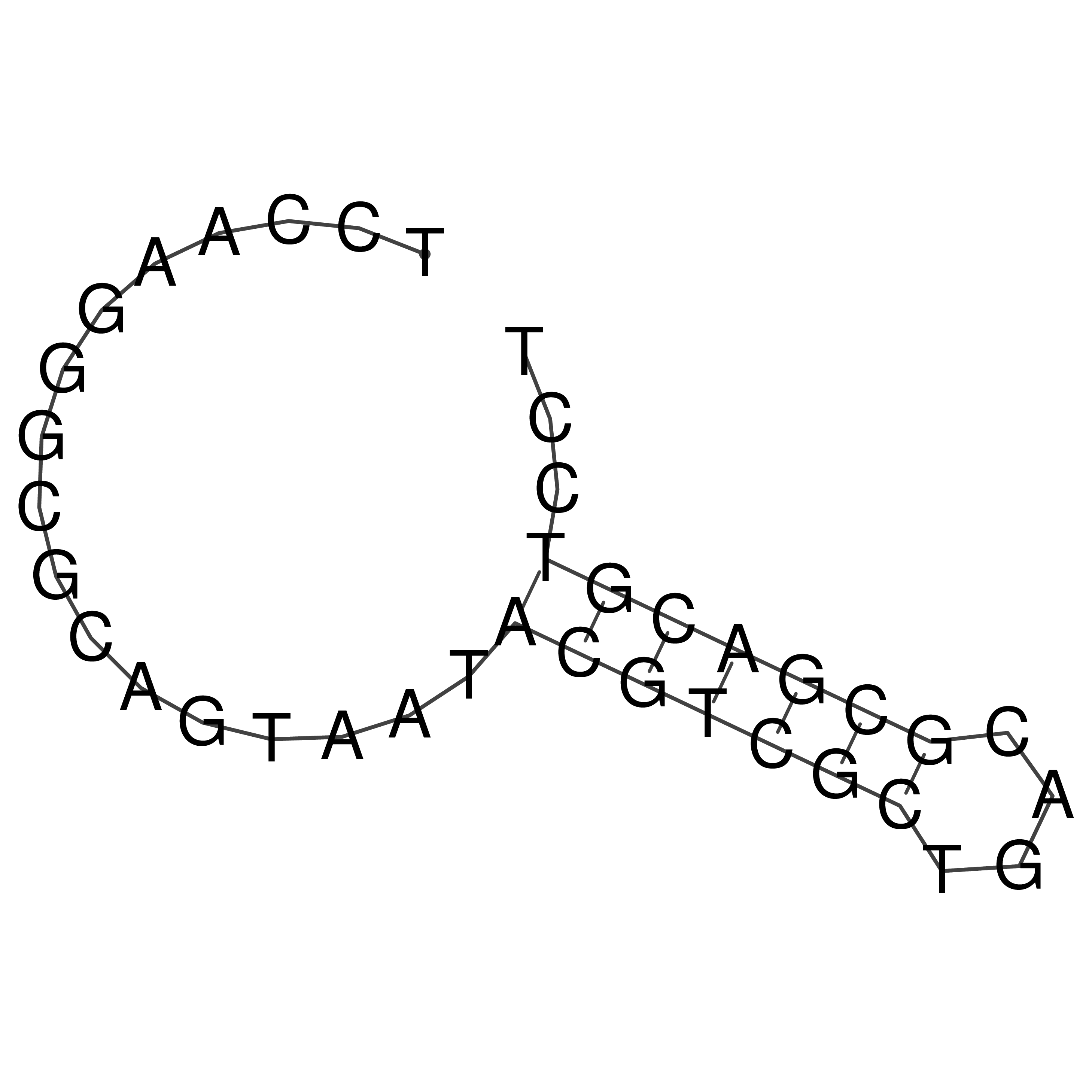
Relaxase
| ID | 4900 | GenBank | WP_280978580 |
| Name | traA_MTX23_RS35295_ISRA436|unnamed |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 123258.33 Da Isoelectric Point: 10.3384
>WP_280978580.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [unclassified Bradyrhizobium]
MAITHFTPQLIGRGDGRSAVLAAAYRHCAKMEFEAEGRTVDYSGKRGMIHEEFQVPADAPAWLRSMIADR
SVAGASEAFWNRVEAFEKRADAQFAKEFIIALPVEQTTEQNIALMRQFVVEQVLSRGQVADWVYHDEPGN
PHVHLMTTLRPLTENGFGPKRVAVIGEDGQPTRMKSGKIAYRLWAGDKQEFLEQRNGWLDLQNQHLAMNG
HEIRVDGRSYVERGIDVVPTTHIGVAAKAIQRRAEREGRDPDLERLRVFDENRAETLRRIQRRPEIVLDM
ISREKSIFDERDVGKLLHRYVDDAGTFHHLMARVLQSPDALRLEGESVDFATAARAPAKYTTRELIRLEA
DMARRAIHLSGSTAFAIKTPVIAAVSARHANLSAEQRSAIEHFASGTRITAVVGRAGAGKTTMMRAAREV
WEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWKSGRDGLNDRTVFVLDEAGMVASRQMALFVE
TASKAGAKLVLVGDPEQLQPIEAGAAFRAIVERIGYAELETIYRQRQQWMRDASLDLARGRTARALSAYR
HHGRLFGSELKAQAIDNLIADWNRDYDPSKSTLILAHLRRDVRALNEMARGKLVERGLVGEGYVFRTEDG
ERRFAAGDQIVFLKNEGSLGVKNGMIGKVVEAAPAFLVAEIGEGDGRRRVEVDQRFYRNIDHGYATTVHK
AQGATVDRVKVLATLSLDKHLTYVALTRHREDVALYYGRRSFQKAGGVIPILSQRRAKETTLDYERGSLY
RDALRFAANRGLHIVNVARTLMRDRLRWTVRQKLRLPEFGRRLRALGEKLGLIDSRKPIMNTAHKEEKPM
VAGVTTFPKSLIDTVEDSILADPAIKKQWEQVSTRFRLVFAAPERAFRATNFDAVLKDSTAATSTLARLA
SNPESFGALRGKTGLLAGKADKQERQRAEANVPALKRDIERYLRLRAEAEHKHEAEERAIRQRVAIDIPA
LSRDARRVLERVRDAIDRNDLPSALEFALADRMVKAEIDGFNKAIAERFGDRTFLPNAAREANGPAFEKA
AAGMDPARRDQLKEAWPAMRAAQQLAAQERSAQALKQAEAQRLTQSQGQTLK
MAITHFTPQLIGRGDGRSAVLAAAYRHCAKMEFEAEGRTVDYSGKRGMIHEEFQVPADAPAWLRSMIADR
SVAGASEAFWNRVEAFEKRADAQFAKEFIIALPVEQTTEQNIALMRQFVVEQVLSRGQVADWVYHDEPGN
PHVHLMTTLRPLTENGFGPKRVAVIGEDGQPTRMKSGKIAYRLWAGDKQEFLEQRNGWLDLQNQHLAMNG
HEIRVDGRSYVERGIDVVPTTHIGVAAKAIQRRAEREGRDPDLERLRVFDENRAETLRRIQRRPEIVLDM
ISREKSIFDERDVGKLLHRYVDDAGTFHHLMARVLQSPDALRLEGESVDFATAARAPAKYTTRELIRLEA
DMARRAIHLSGSTAFAIKTPVIAAVSARHANLSAEQRSAIEHFASGTRITAVVGRAGAGKTTMMRAAREV
WEAAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWKSGRDGLNDRTVFVLDEAGMVASRQMALFVE
TASKAGAKLVLVGDPEQLQPIEAGAAFRAIVERIGYAELETIYRQRQQWMRDASLDLARGRTARALSAYR
HHGRLFGSELKAQAIDNLIADWNRDYDPSKSTLILAHLRRDVRALNEMARGKLVERGLVGEGYVFRTEDG
ERRFAAGDQIVFLKNEGSLGVKNGMIGKVVEAAPAFLVAEIGEGDGRRRVEVDQRFYRNIDHGYATTVHK
AQGATVDRVKVLATLSLDKHLTYVALTRHREDVALYYGRRSFQKAGGVIPILSQRRAKETTLDYERGSLY
RDALRFAANRGLHIVNVARTLMRDRLRWTVRQKLRLPEFGRRLRALGEKLGLIDSRKPIMNTAHKEEKPM
VAGVTTFPKSLIDTVEDSILADPAIKKQWEQVSTRFRLVFAAPERAFRATNFDAVLKDSTAATSTLARLA
SNPESFGALRGKTGLLAGKADKQERQRAEANVPALKRDIERYLRLRAEAEHKHEAEERAIRQRVAIDIPA
LSRDARRVLERVRDAIDRNDLPSALEFALADRMVKAEIDGFNKAIAERFGDRTFLPNAAREANGPAFEKA
AAGMDPARRDQLKEAWPAMRAAQQLAAQERSAQALKQAEAQRLTQSQGQTLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 5103 | GenBank | WP_280978577 |
| Name | traG_MTX23_RS35310_ISRA436|unnamed |
UniProt ID | _ |
| Length | 646 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 646 a.a. Molecular weight: 70108.01 Da Isoelectric Point: 9.4757
>WP_280978577.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [unclassified Bradyrhizobium]
MKANKVALAIAPGVLMAGLCLALTGIEAWLSKFGTTPKAHLVLGRIGIALPYVMAAATGVIFLFAAAGAT
AIRYAGWSVVFGGLLMVAIAASREIARLHSFANAVPAGKELLAYTDPSTVIGATIAALTAVFGFRVAMRG
NAAFAAAEPRRIRGQRAIHGESNWMAITEAARLFPEPGGIVVGERYRVDRDAVADQSFRADDPETWGAGG
KSPLLCFDASFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSNEVAPMVTTHRHGAGRQVFVLD
PRSSEIGFNILDWIGEHGATKEEDIAAVASWIMTDNPRQGSARDDFFRASALQLLTALIADVCLSGHTEK
KDQHLRQVRANLSEPEPKLRQRLQTIYDNSESGFVKENVAPFIAMTPETFSGVYANAVKETHWLSYANYA
ALVSGSSFTTAELAEGRTDVFINLDLKTLENHAGLARVIIGSLLNAIYNRNGEANGRTLFLLDEVARLGY
LRILETARDAGRKYGITLLMLYQSIGQMREAYGGRDATSKWFESASWISFSAINDPETADYISKRCGDTT
VEVDQLSRSSQMSGSSRTRSKQLARRPLMLPHEVLRMRTDEQIIFTAGNPPLRCGRAIWFRRADMRTCVG
ENRFHRREAGGRSRRS
MKANKVALAIAPGVLMAGLCLALTGIEAWLSKFGTTPKAHLVLGRIGIALPYVMAAATGVIFLFAAAGAT
AIRYAGWSVVFGGLLMVAIAASREIARLHSFANAVPAGKELLAYTDPSTVIGATIAALTAVFGFRVAMRG
NAAFAAAEPRRIRGQRAIHGESNWMAITEAARLFPEPGGIVVGERYRVDRDAVADQSFRADDPETWGAGG
KSPLLCFDASFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSNEVAPMVTTHRHGAGRQVFVLD
PRSSEIGFNILDWIGEHGATKEEDIAAVASWIMTDNPRQGSARDDFFRASALQLLTALIADVCLSGHTEK
KDQHLRQVRANLSEPEPKLRQRLQTIYDNSESGFVKENVAPFIAMTPETFSGVYANAVKETHWLSYANYA
ALVSGSSFTTAELAEGRTDVFINLDLKTLENHAGLARVIIGSLLNAIYNRNGEANGRTLFLLDEVARLGY
LRILETARDAGRKYGITLLMLYQSIGQMREAYGGRDATSKWFESASWISFSAINDPETADYISKRCGDTT
VEVDQLSRSSQMSGSSRTRSKQLARRPLMLPHEVLRMRTDEQIIFTAGNPPLRCGRAIWFRRADMRTCVG
ENRFHRREAGGRSRRS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5104 | GenBank | WP_280978493 |
| Name | t4cp2_MTX23_RS35820_ISRA436|unnamed |
UniProt ID | _ |
| Length | 817 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 817 a.a. Molecular weight: 91343.09 Da Isoelectric Point: 6.3242
>WP_280978493.1 MULTISPECIES: conjugal transfer protein TrbE [unclassified Bradyrhizobium]
MVALRSFRHSGPSFADLVPYAGLVANGIVLLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINTILARLG
SGWMIQVEAVRVPTTDYPARRDCHFPDPVTRAIDDERRGRFEQERGHFESRHAIILTWRPPERRRAGLAR
YIYSDVESRSASYADTAIESFRTSIREVEQYLASVLTIRRMETREIEERDGTRMARYDELFQFIRFCITG
ENHPIRLPDIPMYLDWLATAELQHGLSPTVENRFLGVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQDARQKLERTRKKWQQKVRPFIDQLFQTQSRSIDHDAAAMVAEAEDAIAEASSQLAAYGYYTPVLVL
FDESSARLQEKAEAVRRLIQAEGFGARIETLNATEAFLGSLPGNWYANIREPLISTRNLADLIPLNSVWS
GNPIAPCPFYPPSSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYAGAQIFA
FDKGRSMLPLTLAAGGDHYEIGGDDAGGAAALAFCPLSDLLTDTDRAWAAEWIETLVAVQGVVITPDHRN
AISRQVGLMAEARGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAERDGLQLGSFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLSGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAVPKREYYVASPEGRRLFNMALGP
VALSFVGASGKEDLKRILALKKGKANWPVHWLKERGIGHAETLLADF
MVALRSFRHSGPSFADLVPYAGLVANGIVLLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINTILARLG
SGWMIQVEAVRVPTTDYPARRDCHFPDPVTRAIDDERRGRFEQERGHFESRHAIILTWRPPERRRAGLAR
YIYSDVESRSASYADTAIESFRTSIREVEQYLASVLTIRRMETREIEERDGTRMARYDELFQFIRFCITG
ENHPIRLPDIPMYLDWLATAELQHGLSPTVENRFLGVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQDARQKLERTRKKWQQKVRPFIDQLFQTQSRSIDHDAAAMVAEAEDAIAEASSQLAAYGYYTPVLVL
FDESSARLQEKAEAVRRLIQAEGFGARIETLNATEAFLGSLPGNWYANIREPLISTRNLADLIPLNSVWS
GNPIAPCPFYPPSSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLALIAAQFRRYAGAQIFA
FDKGRSMLPLTLAAGGDHYEIGGDDAGGAAALAFCPLSDLLTDTDRAWAAEWIETLVAVQGVVITPDHRN
AISRQVGLMAEARGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAERDGLQLGSFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLSGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVLLATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAVPKREYYVASPEGRRLFNMALGP
VALSFVGASGKEDLKRILALKKGKANWPVHWLKERGIGHAETLLADF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 189740..199311
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MTX23_RS35785 (MTX23_35695) | 185262..186296 | - | 1035 | WP_280978499 | plasmid partitioning protein RepB | - |
| MTX23_RS35790 (MTX23_35700) | 186293..187507 | - | 1215 | WP_280978498 | plasmid partitioning protein RepA | - |
| MTX23_RS35795 (MTX23_35705) | 188111..188542 | + | 432 | WP_280978497 | hypothetical protein | - |
| MTX23_RS35800 (MTX23_35710) | 189099..189722 | + | 624 | WP_280978496 | acyl-homoserine-lactone synthase TraI | - |
| MTX23_RS35805 (MTX23_35715) | 189740..190708 | + | 969 | WP_280978495 | P-type conjugative transfer ATPase TrbB | virB11 |
| MTX23_RS35810 (MTX23_35720) | 190698..191078 | + | 381 | WP_280978494 | TrbC/VirB2 family protein | virB2 |
| MTX23_RS35815 (MTX23_35725) | 191071..191370 | + | 300 | WP_027584282 | conjugal transfer protein TrbD | virB3 |
| MTX23_RS35820 (MTX23_35730) | 191380..193833 | + | 2454 | WP_280978493 | conjugal transfer protein TrbE | virb4 |
| MTX23_RS35825 (MTX23_35735) | 193805..194611 | + | 807 | WP_280978492 | P-type conjugative transfer protein TrbJ | virB5 |
| MTX23_RS35830 (MTX23_35740) | 194608..194796 | + | 189 | WP_280978491 | entry exclusion protein TrbK | - |
| MTX23_RS35835 (MTX23_35745) | 194814..195974 | + | 1161 | WP_280990759 | P-type conjugative transfer protein TrbL | virB6 |
| MTX23_RS35840 (MTX23_35750) | 196018..196680 | + | 663 | WP_280978490 | conjugal transfer protein TrbF | virB8 |
| MTX23_RS35845 (MTX23_35755) | 196698..197537 | + | 840 | WP_280978489 | P-type conjugative transfer protein TrbG | virB9 |
| MTX23_RS35850 (MTX23_35760) | 197540..197992 | + | 453 | WP_280978488 | conjugal transfer protein TrbH | - |
| MTX23_RS35855 (MTX23_35765) | 198004..199311 | + | 1308 | WP_280978487 | IncP-type conjugal transfer protein TrbI | virB10 |
| MTX23_RS35860 (MTX23_35770) | 200361..200627 | - | 267 | WP_280978486 | hypothetical protein | - |
| MTX23_RS35865 (MTX23_35775) | 201104..202009 | + | 906 | WP_280978485 | LysR family transcriptional regulator | - |
| MTX23_RS35870 (MTX23_35780) | 202091..203863 | - | 1773 | WP_280978484 | GMC family oxidoreductase N-terminal domain-containing protein | - |
Host bacterium
| ID | 7785 | GenBank | NZ_CP094520 |
| Plasmid name | ISRA436|unnamed | Incompatibility group | - |
| Plasmid size | 316663 bp | Coordinate of oriT [Strand] | 84507..84544 [-] |
| Host baterium | Bradyrhizobium sp. ISRA436 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |