Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107305 |
| Name | oriT_pYS1 |
| Organism | Burkholderia cepacia |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_019369 (29988..30084 [+], 97 nt) |
| oriT length | 97 nt |
| IRs (inverted repeats) | 71..76, 80..85 (GGCGTT..AACGCC) 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 97 nt
>oriT_pYS1
GAATAAGGGGGAGCGAAGATAGATAACCGGCTTGCCGGTTAGCTAACTTCGCCCATCTTGCCCGCCTTACGGCGTTGATAACGCCAAGGAAAGTCTA
GAATAAGGGGGAGCGAAGATAGATAACCGGCTTGCCGGTTAGCTAACTTCGCCCATCTTGCCCGCCTTACGGCGTTGATAACGCCAAGGAAAGTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file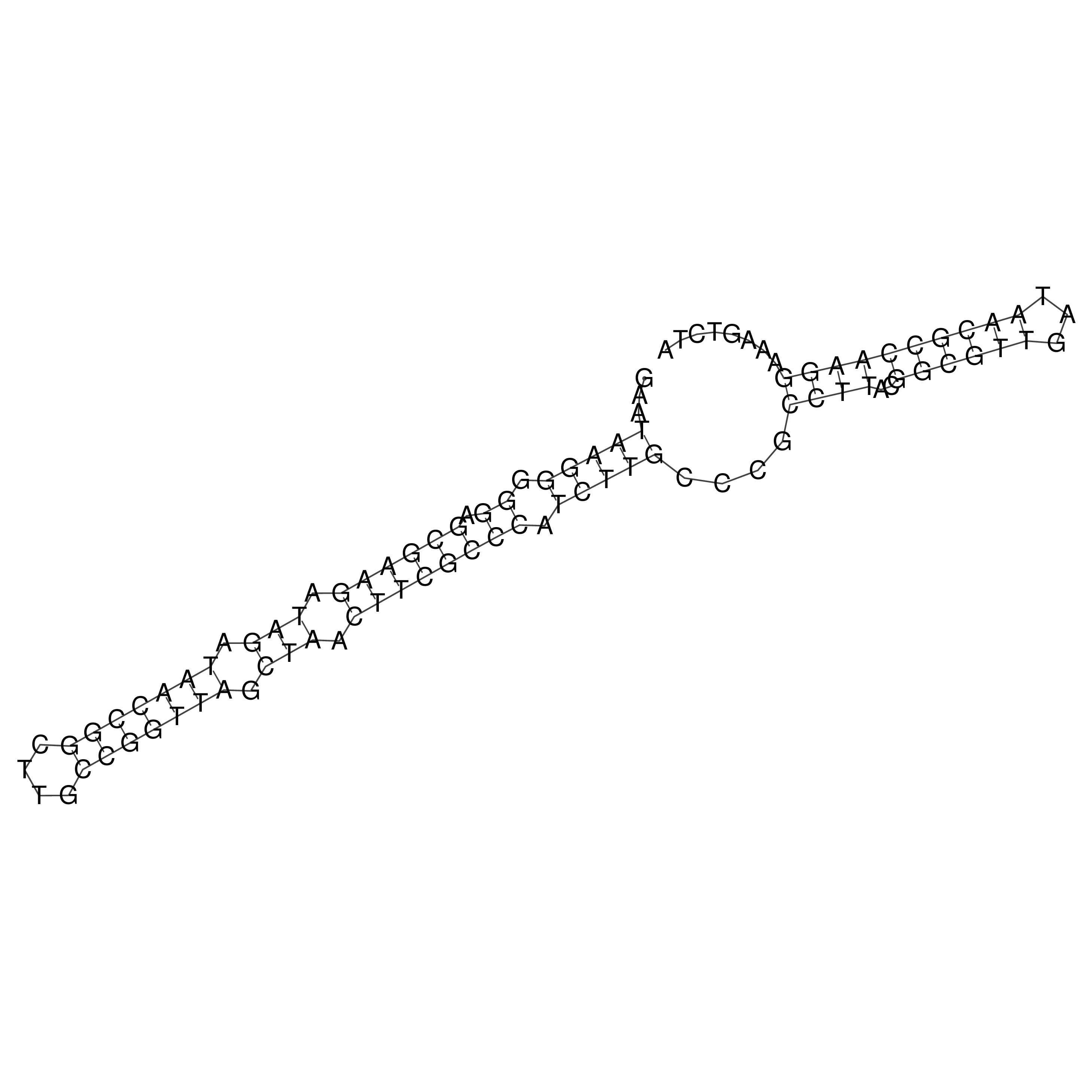
Relaxase
| ID | 4859 | GenBank | WP_015063387 |
| Name | Relaxase_HTB02_RS00140_pYS1 |
UniProt ID | J9R8K8 |
| Length | 747 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 747 a.a. Molecular weight: 82577.51 Da Isoelectric Point: 10.8056
>WP_015063387.1 TraI/MobA(P) family conjugative relaxase [Burkholderia cepacia]
MILKHVPMRSLGKSDFAGLVNYVTDSQSKEHRLGVVQITNCEAGSVRDAITEVLATQHTNTRAIGDKTYH
LLIGFPPGENPSADTLKQIEERVCAALGYGEHQRVSAVHNDTDHLHIHLAINKIHPTRHTMHEPFQAYRT
MAEIAAKLEEEYGLQRVNHESRRSLAEGRAGDMEQHSGIESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRAQALEQARRRKDRLVEAAKRSNRLRRATIKVVGENRTNKKLLYAQASKALRDE
IQAINKQYQKDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLQGNTIQGRGEARPGRAPV
TDNITKKGTIIFRAGLSAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFKAQVIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAAAHDDAARPVAVA
RAGFGRTDAAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHLEQQGT
QPDHQLRRGISRPGTGVAAAPPGLAAADKYIAEREAKRSKGFDIPKHARYTAGDGALAFQGVRNIEGQAL
ALLKRGDEVMVMPIDQATARRLSRIAVGDAVTITPKGSIKTSKGRSR
MILKHVPMRSLGKSDFAGLVNYVTDSQSKEHRLGVVQITNCEAGSVRDAITEVLATQHTNTRAIGDKTYH
LLIGFPPGENPSADTLKQIEERVCAALGYGEHQRVSAVHNDTDHLHIHLAINKIHPTRHTMHEPFQAYRT
MAEIAAKLEEEYGLQRVNHESRRSLAEGRAGDMEQHSGIESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRAQALEQARRRKDRLVEAAKRSNRLRRATIKVVGENRTNKKLLYAQASKALRDE
IQAINKQYQKDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLQGNTIQGRGEARPGRAPV
TDNITKKGTIIFRAGLSAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFKAQVIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAAAHDDAARPVAVA
RAGFGRTDAAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHLEQQGT
QPDHQLRRGISRPGTGVAAAPPGLAAADKYIAEREAKRSKGFDIPKHARYTAGDGALAFQGVRNIEGQAL
ALLKRGDEVMVMPIDQATARRLSRIAVGDAVTITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | J9R8K8 |
T4CP
| ID | 5042 | GenBank | WP_015063366 |
| Name | t4cp2_HTB02_RS00035_pYS1 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 94175.10 Da Isoelectric Point: 6.3996
>WP_015063366.1 VirB4 family type IV secretion/conjugal transfer ATPase [Burkholderia cepacia]
MIEAIAIGIAVLGAVLLLILFSRIRQVDAELKLKKHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMASWL
YKGDDNASSTDEQREMVSFRINQALAGLGSGWMVHVDAVRRPAPNYSERGVSTFTDPVSAAIDEERRRLF
ESLGTMYEGHFVFTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTVGLIEQFKREIRSLESRLSSAVKLT
RLQGHKMATEEGREVTHDDFLSWLQFCVTGNYHPVQLPSNPMYLDAVIGGQELWGGVVPKVGRKFIQVVS
IEGFPLESTPGLLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSVNQD
AATMVADAEAAIAEVNSGLVAAGYFTSVVVLMDEDRERLAASALSVEKAINRLAFAARIETINTMDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLAPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAAINAGSKGKTGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVDTTPAQRNEIGHAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERGLKGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNKRQIEILATAVPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLQAKFGHEWVDHWLAG
RNLNLNDYLEAA
MIEAIAIGIAVLGAVLLLILFSRIRQVDAELKLKKHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMASWL
YKGDDNASSTDEQREMVSFRINQALAGLGSGWMVHVDAVRRPAPNYSERGVSTFTDPVSAAIDEERRRLF
ESLGTMYEGHFVFTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTVGLIEQFKREIRSLESRLSSAVKLT
RLQGHKMATEEGREVTHDDFLSWLQFCVTGNYHPVQLPSNPMYLDAVIGGQELWGGVVPKVGRKFIQVVS
IEGFPLESTPGLLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSVNQD
AATMVADAEAAIAEVNSGLVAAGYFTSVVVLMDEDRERLAASALSVEKAINRLAFAARIETINTMDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLAPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAAINAGSKGKTGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVDTTPAQRNEIGHAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERGLKGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNKRQIEILATAVPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLQAKFGHEWVDHWLAG
RNLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 5043 | GenBank | WP_015063386 |
| Name | t4cp2_HTB02_RS00135_pYS1 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69541.54 Da Isoelectric Point: 8.5000
>WP_015063386.1 type IV secretory system conjugative DNA transfer family protein [Burkholderia cepacia]
MKIKMNNAVGPQVRSAKAKTPKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPAATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNEGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKASDRLRQAVEAGE
GITI
MKIKMNNAVGPQVRSAKAKTPKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPAATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNEGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARAAIPAPKASDRLRQAVEAGE
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2390..13036
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HTB02_RS00005 (pYS10001) | 1..1224 | - | 1224 | WP_015063361 | plasmid replication initiator TrfA | - |
| HTB02_RS00010 (pYS10002) | 1272..1610 | - | 339 | WP_011304114 | single-stranded DNA-binding protein | - |
| HTB02_RS00015 (pYS10003) | 1719..2078 | + | 360 | WP_015063362 | transcriptional regulator | - |
| HTB02_RS00020 (pYS10004) | 2390..3355 | + | 966 | WP_015063363 | P-type conjugative transfer ATPase TrbB | virB11 |
| HTB02_RS00025 (pYS10005) | 3377..3841 | + | 465 | WP_015063364 | conjugal transfer system pilin TrbC | virB2 |
| HTB02_RS00030 (pYS10006) | 3845..4156 | + | 312 | WP_015063365 | conjugal transfer protein TrbD | virB3 |
| HTB02_RS00035 (pYS10007) | 4153..6711 | + | 2559 | WP_015063366 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HTB02_RS00040 (pYS10008) | 6708..7496 | + | 789 | WP_015063367 | conjugal transfer protein TrbF | virB8 |
| HTB02_RS00045 (pYS10009) | 7514..8407 | + | 894 | WP_015063368 | P-type conjugative transfer protein TrbG | virB9 |
| HTB02_RS00050 (pYS10010) | 8410..8871 | + | 462 | WP_015063369 | lipoprotein | - |
| HTB02_RS00055 (pYS10011) | 8876..10267 | + | 1392 | WP_015063370 | TrbI/VirB10 family protein | virB10 |
| HTB02_RS00060 (pYS10012) | 10284..11057 | + | 774 | WP_015063371 | P-type conjugative transfer protein TrbJ | virB5 |
| HTB02_RS00065 (pYS10013) | 11067..11288 | + | 222 | WP_011304125 | entry exclusion lipoprotein TrbK | - |
| HTB02_RS00070 (pYS10014) | 11300..13036 | + | 1737 | WP_015063373 | P-type conjugative transfer protein TrbL | virB6 |
| HTB02_RS00075 (pYS10015) | 13054..13632 | + | 579 | WP_015063374 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| HTB02_RS00080 (pYS10016) | 13634..14269 | + | 636 | WP_015063375 | transglycosylase SLT domain-containing protein | - |
| HTB02_RS00085 (pYS10017) | 14299..14568 | + | 270 | WP_015063376 | hypothetical protein | - |
| HTB02_RS00090 (pYS10018) | 14565..15263 | + | 699 | WP_015063377 | TraX family protein | - |
| HTB02_RS00095 (pYS10019) | 15279..15722 | + | 444 | WP_015063378 | OmpA family protein | - |
| HTB02_RS00100 (pYS10020) | 15914..16588 | + | 675 | WP_015063379 | class I SAM-dependent methyltransferase | - |
| HTB02_RS00105 (pYS10022) | 16679..17188 | + | 510 | WP_181411585 | hypothetical protein | - |
| HTB02_RS00110 (pYS10021) | 17145..17813 | - | 669 | WP_015063381 | recombinase family protein | - |
Host bacterium
| ID | 7741 | GenBank | NC_019369 |
| Plasmid name | pYS1 | Incompatibility group | - |
| Plasmid size | 82988 bp | Coordinate of oriT [Strand] | 29988..30084 [+] |
| Host baterium | Burkholderia cepacia |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |