Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107039 |
| Name | oriT_p334D3 |
| Organism | Enterobacter hormaechei strain 334D3 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP119681 (2157..2255 [+], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..30, 35..41 (TAACCGG..CCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_p334D3
GAATAAGGGACAGTGAAGATAGATAACCGGCTCACCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
GAATAAGGGACAGTGAAGATAGATAACCGGCTCACCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file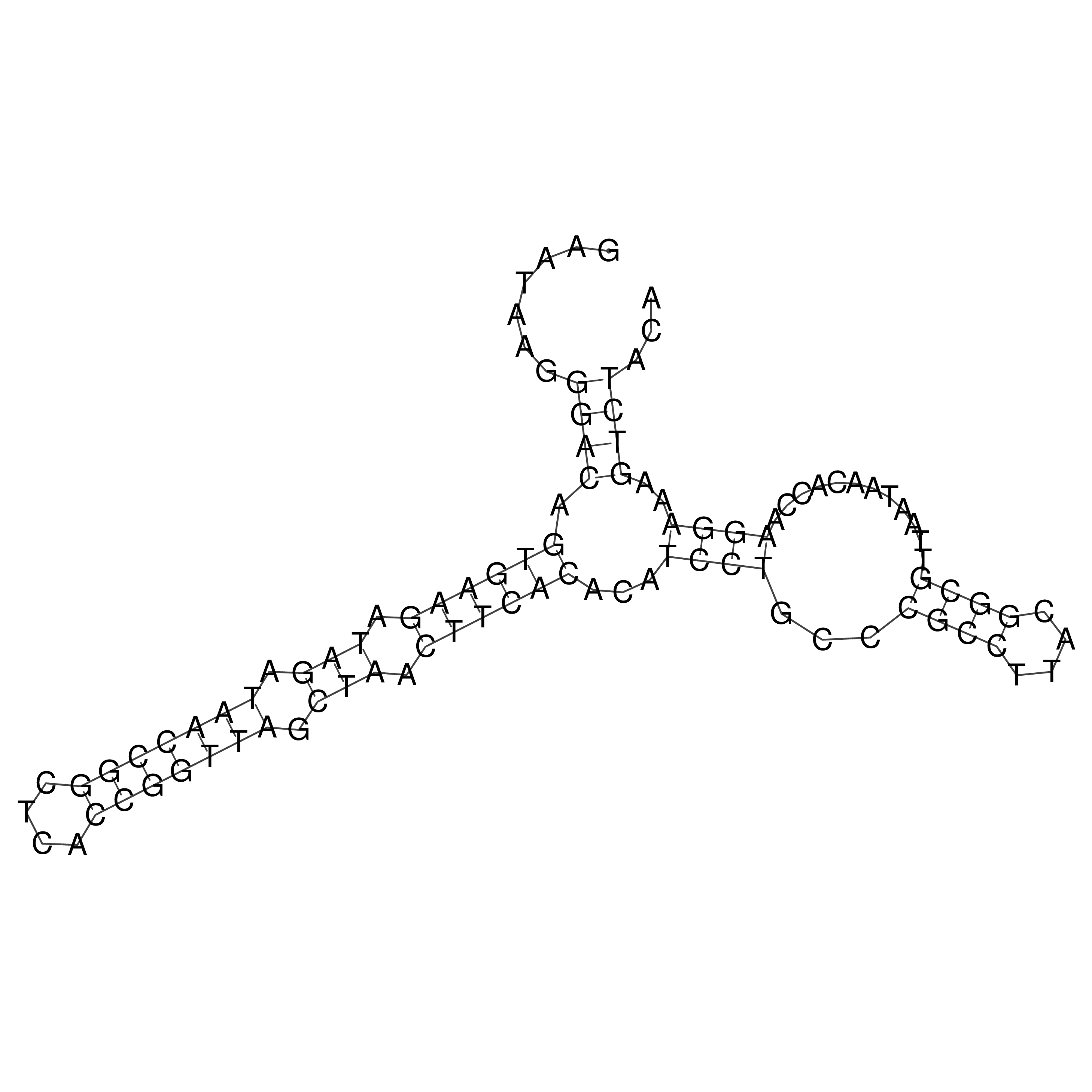
Relaxase
| ID | 4648 | GenBank | WP_277376365 |
| Name | Relaxase_P3J69_RS00005_p334D3 |
UniProt ID | _ |
| Length | 746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 82022.65 Da Isoelectric Point: 10.7900
>WP_277376365.1 TraI/MobA(P) family conjugative relaxase [Enterobacter hormaechei]
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTAAKRQYRKDPIRLRVNTV
ELYAKYKAEQQTLTTARGQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRTNKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGSADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTAAKRQYRKDPIRLRVNTV
ELYAKYKAEQQTLTTARGQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRTNKKLLYAQASKALRSE
IQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGSADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4749 | GenBank | WP_277376357 |
| Name | t4cp2_P3J69_RS00135_p334D3 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93996.15 Da Isoelectric Point: 6.3901
>WP_277376357.1 VirB4 family type IV secretion/conjugal transfer ATPase [Enterobacter hormaechei]
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLISQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLISQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4750 | GenBank | WP_000694765 |
| Name | t4cp2_P3J69_RS00310_p334D3 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69897.09 Da Isoelectric Point: 8.2993
>WP_000694765.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Bacteria]
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 16973..27662
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| P3J69_RS00090 (P3J69_00090) | 12060..12488 | - | 429 | WP_277376358 | antirestriction protein | - |
| P3J69_RS00095 (P3J69_00095) | 12657..12926 | + | 270 | WP_001260984 | type II toxin-antitoxin system ParD family antitoxin | - |
| P3J69_RS00100 (P3J69_00100) | 12923..13237 | + | 315 | WP_000119407 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| P3J69_RS00105 (P3J69_00105) | 14579..15799 | - | 1221 | WP_001178704 | plasmid replication initiator TrfA | - |
| P3J69_RS00110 (P3J69_00110) | 15846..16187 | - | 342 | WP_000019167 | single-stranded DNA-binding protein | - |
| P3J69_RS00115 (P3J69_00115) | 16301..16663 | + | 363 | WP_000283041 | transcriptional regulator | - |
| P3J69_RS00120 (P3J69_00120) | 16973..17935 | + | 963 | WP_031942719 | P-type conjugative transfer ATPase TrbB | virB11 |
| P3J69_RS00125 (P3J69_00125) | 17952..18416 | + | 465 | WP_001149042 | conjugal transfer system pilin TrbC | virB2 |
| P3J69_RS00130 (P3J69_00130) | 18420..18731 | + | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| P3J69_RS00135 (P3J69_00135) | 18728..21286 | + | 2559 | WP_277376357 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| P3J69_RS00140 (P3J69_00140) | 21283..22065 | + | 783 | WP_031942721 | conjugal transfer protein TrbF | virB8 |
| P3J69_RS00145 (P3J69_00145) | 22083..22982 | + | 900 | WP_000753228 | P-type conjugative transfer protein TrbG | virB9 |
| P3J69_RS00150 (P3J69_00150) | 22985..23476 | + | 492 | WP_001227154 | conjugal transfer protein TrbH | - |
| P3J69_RS00155 (P3J69_00155) | 23481..24902 | + | 1422 | WP_000138182 | TrbI/VirB10 family protein | virB10 |
| P3J69_RS00160 (P3J69_00160) | 24920..25684 | + | 765 | WP_000802794 | P-type conjugative transfer protein TrbJ | virB5 |
| P3J69_RS00165 (P3J69_00165) | 25694..25921 | + | 228 | WP_000644148 | entry exclusion lipoprotein TrbK | - |
| P3J69_RS00170 (P3J69_00170) | 25932..27662 | + | 1731 | WP_277376354 | P-type conjugative transfer protein TrbL | virB6 |
| P3J69_RS00175 (P3J69_00175) | 27680..28264 | + | 585 | WP_031942724 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| P3J69_RS00180 (P3J69_00180) | 28278..28922 | + | 645 | WP_031942725 | transglycosylase SLT domain-containing protein | - |
| P3J69_RS00185 (P3J69_00185) | 28919..29185 | + | 267 | WP_000222514 | hypothetical protein | - |
| P3J69_RS00190 (P3J69_00190) | 29262..29897 | + | 636 | WP_280956581 | TraX family protein | - |
| P3J69_RS00195 (P3J69_00195) | 29913..30344 | + | 432 | WP_031942727 | OmpA family protein | - |
| P3J69_RS00200 (P3J69_00200) | 30500..31174 | + | 675 | WP_001270224 | hypothetical protein | - |
| P3J69_RS00205 (P3J69_00205) | 31176..32027 | - | 852 | WP_280956579 | recombinase family protein | - |
Host bacterium
| ID | 7475 | GenBank | NZ_CP119681 |
| Plasmid name | p334D3 | Incompatibility group | IncP1 |
| Plasmid size | 52007 bp | Coordinate of oriT [Strand] | 2157..2255 [+] |
| Host baterium | Enterobacter hormaechei strain 334D3 |
Cargo genes
| Drug resistance gene | blaIMP-13, fosE, sul1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |