Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 107036 |
| Name | oriT1_pHS08-175-2-52445bp-repAIncN |
| Organism | Enterobacter hormaechei subsp. steigerwaltii strain 08-175 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MT010563 (9295..9354 [+], 60 nt) |
| oriT length | 60 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 60 nt
>oriT1_pHS08-175-2-52445bp-repAIncN
GGGTGTCGGGGCGCAGCCCTGACCCAGTCACATAGCGATAGCGGAGTGTATACTGGCTTA
GGGTGTCGGGGCGCAGCCCTGACCCAGTCACATAGCGATAGCGGAGTGTATACTGGCTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file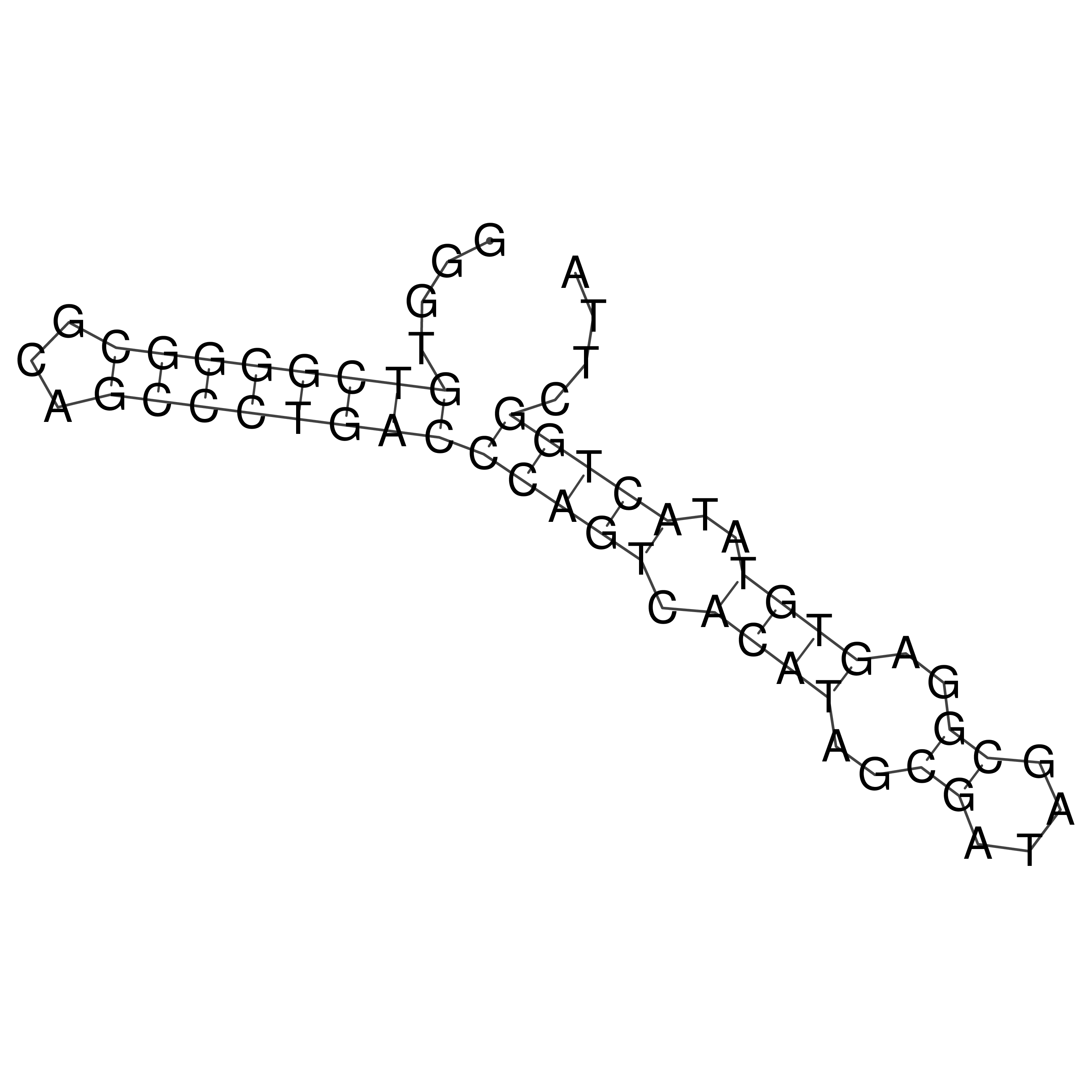
Relaxase
| ID | 4646 | GenBank | WP_000884351 |
| Name | mobF_MJ822_RS00255_pHS08-175-2-52445bp-repAIncN |
UniProt ID | D7RVP6 |
| Length | 1076 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1076 a.a. Molecular weight: 119897.16 Da Isoelectric Point: 6.6071
>WP_000884351.1 MULTISPECIES: MobF family relaxase [Enterobacterales]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLAQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
ISGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D7RVP6 |
Auxiliary protein
| ID | 2075 | GenBank | WP_013188476 |
| Name | WP_013188476_pHS08-175-2-52445bp-repAIncN |
UniProt ID | D7RVK1 |
| Length | 138 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 138 a.a. Molecular weight: 15332.60 Da Isoelectric Point: 5.9670
>WP_013188476.1 MULTISPECIES: hypothetical protein [Enterobacterales]
MPIITAKVSDELLAYIDLVSGGNRSDYLRRCLEAGPGDRESGLKIVADRLSDVNRKLDYIFDRASDADFG
PLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIAEPGKTKAAKAEVERNGYKVWEPKKER
MPIITAKVSDELLAYIDLVSGGNRSDYLRRCLEAGPGDRESGLKIVADRLSDVNRKLDYIFDRASDADFG
PLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIAEPGKTKAAKAEVERNGYKVWEPKKER
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | D7RVK1 |
T4CP
| ID | 4745 | GenBank | WP_000342688 |
| Name | t4cp2_MJ822_RS00260_pHS08-175-2-52445bp-repAIncN |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57762.87 Da Isoelectric Point: 9.6551
>WP_000342688.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVRARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELREI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 12976..22264
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MJ822_RS00355 | 7985..8683 | - | 699 | Protein_9 | relaxase/mobilization nuclease domain-containing protein | - |
| MJ822_RS00055 | 8625..8948 | - | 324 | WP_040090312 | MobC family plasmid mobilization relaxosome protein | - |
| MJ822_RS00060 | 9013..9204 | + | 192 | WP_000165985 | Rop family plasmid primer RNA-binding protein | - |
| MJ822_RS00065 | 9874..9987 | + | 114 | WP_071699851 | replication initiation protein | - |
| MJ822_RS00070 | 10205..10345 | - | 141 | WP_072020761 | colicin R system lysis protein | - |
| MJ822_RS00075 | 10465..10989 | + | 525 | WP_040090309 | colicin R immunity protein | - |
| MJ822_RS00080 | 11017..11838 | - | 822 | Protein_15 | pore-forming bacteriocin colicin R | - |
| MJ822_RS00085 | 11882..12586 | - | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| MJ822_RS00090 | 12650..12934 | - | 285 | Protein_17 | conjugal transfer protein TraG | - |
| MJ822_RS00095 | 12976..14136 | - | 1161 | WP_000101712 | type IV secretion system protein VirB10 | virB10 |
| MJ822_RS00100 | 14136..15020 | - | 885 | WP_000735067 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| MJ822_RS00105 | 15031..15729 | - | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| MJ822_RS00110 | 15719..15877 | - | 159 | WP_012561180 | hypothetical protein | - |
| MJ822_RS00115 | 15948..16988 | - | 1041 | WP_000886022 | type IV secretion system protein | virB6 |
| MJ822_RS00120 | 17004..17231 | - | 228 | WP_000734973 | IncN-type entry exclusion lipoprotein EexN | - |
| MJ822_RS00125 | 17239..17952 | - | 714 | WP_000749362 | type IV secretion system protein | virB5 |
| MJ822_RS00130 | 17970..20570 | - | 2601 | WP_001200711 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| MJ822_RS00135 | 20570..20887 | - | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| MJ822_RS00140 | 20937..21230 | - | 294 | WP_000209137 | TrbC/VirB2 family protein | virB2 |
| MJ822_RS00145 | 21240..21521 | - | 282 | WP_000440698 | transcriptional repressor KorA | - |
| MJ822_RS00150 | 21530..22264 | - | 735 | WP_000033783 | lytic transglycosylase domain-containing protein | virB1 |
| MJ822_RS00155 | 22307..22678 | + | 372 | WP_011867773 | H-NS family nucleoid-associated regulatory protein | - |
| MJ822_RS00160 | 22694..23071 | + | 378 | WP_000504250 | hypothetical protein | - |
| MJ822_RS00165 | 23034..23348 | + | 315 | WP_001749965 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| MJ822_RS00170 | 23462..23695 | + | 234 | WP_001191790 | hypothetical protein | - |
| MJ822_RS00175 | 23745..24392 | + | 648 | WP_001532074 | restriction endonuclease | - |
| MJ822_RS00180 | 24397..24594 | + | 198 | WP_013188474 | hypothetical protein | - |
| MJ822_RS00185 | 24614..24885 | - | 272 | Protein_36 | IS1 family transposase | - |
| MJ822_RS00190 | 24979..26412 | + | 1434 | WP_001288433 | DNA cytosine methyltransferase | - |
Host bacterium
| ID | 7472 | GenBank | NZ_MT010563 |
| Plasmid name | pHS08-175-2-52445bp-repAIncN | Incompatibility group | IncN |
| Plasmid size | 52445 bp | Coordinate of oriT [Strand] | 9295..9354 [+]; 40830..40930 [+] |
| Host baterium | Enterobacter hormaechei subsp. steigerwaltii strain 08-175 |
Cargo genes
| Drug resistance gene | fosA3, blaCTX-M-125 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |