Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106974 |
| Name | oriT_p50821_KPC |
| Organism | Morganella morganii strain MM50821 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP070550 (1318..1417 [-], 100 nt) |
| oriT length | 100 nt |
| IRs (inverted repeats) | 78..83, 90..95 (AAAAAA..TTTTTT) 78..83, 89..94 (AAAAAA..TTTTTT) 32..39, 42..49 (AGCGTGAT..ATCACGCT) 18..24, 36..42 (TAAATCA..TGATTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 100 nt
>oriT_p50821_KPC
TTTGTTTTTTTTTCTTTTAAATCAGTGCGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTTGGTAG
TTTGTTTTTTTTTCTTTTAAATCAGTGCGATAGCGTGATTTATCACGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file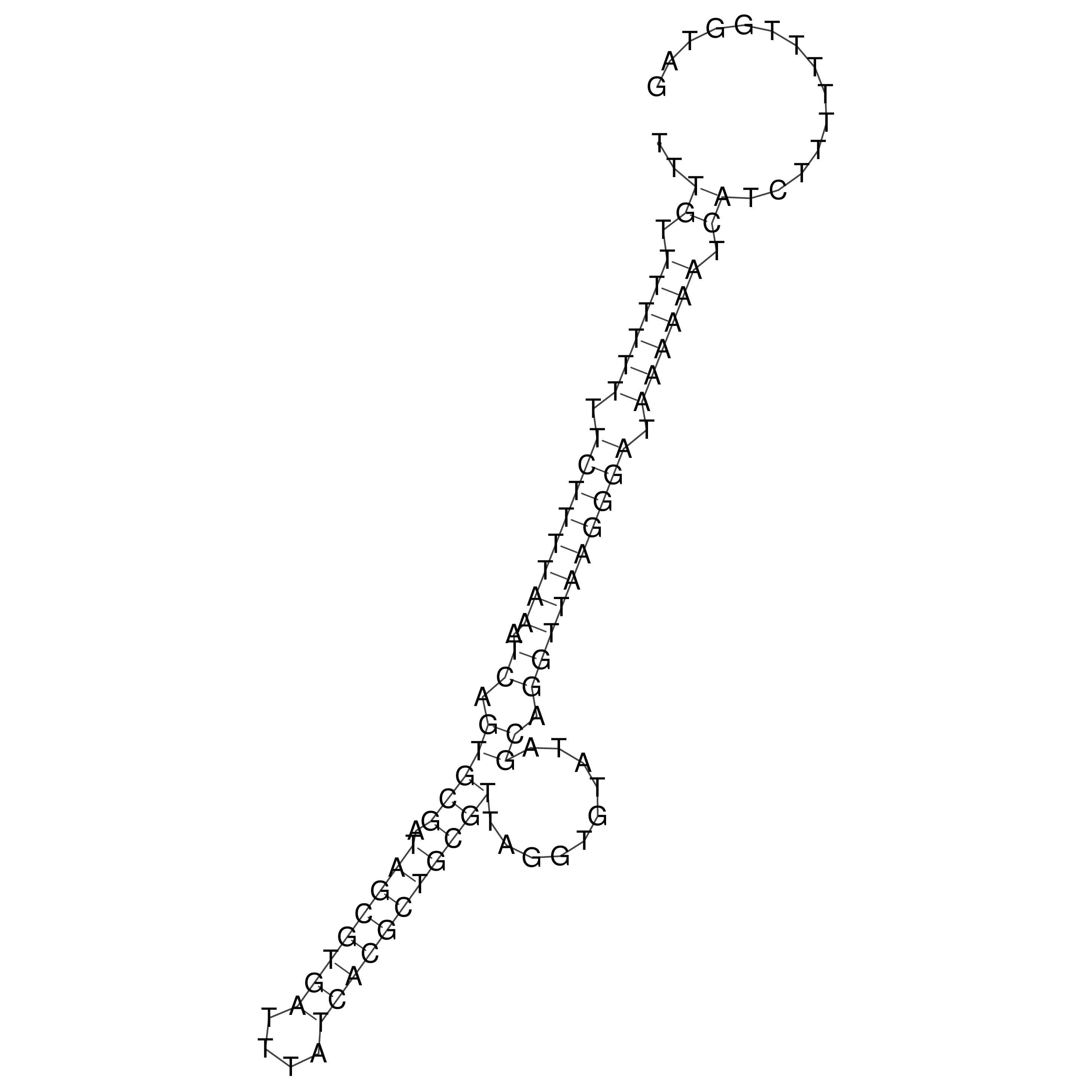
Relaxase
| ID | 4605 | GenBank | WP_000130000 |
| Name | Replic_Relax_JW297_RS00070_p50821_KPC |
UniProt ID | R4WML4 |
| Length | 101 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 101 a.a. Molecular weight: 11477.11 Da Isoelectric Point: 7.5204
>WP_000130000.1 MULTISPECIES: PadR family transcriptional regulator [Pseudomonadota]
MTDKDLYGGLIRLHILHHAAEEPVFGLGIIEELRRHGYEMSAGTVYPMLHGLEKKGYLTSRHERTGRRER
RVYDITEQGRTALADAKTKVKELFGELVEGG
MTDKDLYGGLIRLHILHHAAEEPVFGLGIIEELRRHGYEMSAGTVYPMLHGLEKKGYLTSRHERTGRRER
RVYDITEQGRTALADAKTKVKELFGELVEGG
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | R4WML4 |
Host bacterium
| ID | 7411 | GenBank | NZ_CP070550 |
| Plasmid name | p50821_KPC | Incompatibility group | IncR |
| Plasmid size | 54347 bp | Coordinate of oriT [Strand] | 1318..1417 [-] |
| Host baterium | Morganella morganii strain MM50821 |
Cargo genes
| Drug resistance gene | catB3, ARR-3, qacE, sul1, mph(A), blaKPC-2, aac(6')-Ib |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |