Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106952 |
| Name | oriT_CIP 109850|unnamed |
| Organism | Sinorhizobium numidicum strain CIP 109850 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP120369 (628761..628800 [-], 40 nt) |
| oriT length | 40 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 40 nt
TCCAAGGGCGCAATTATACGTCGCTGCCGCGACGTCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file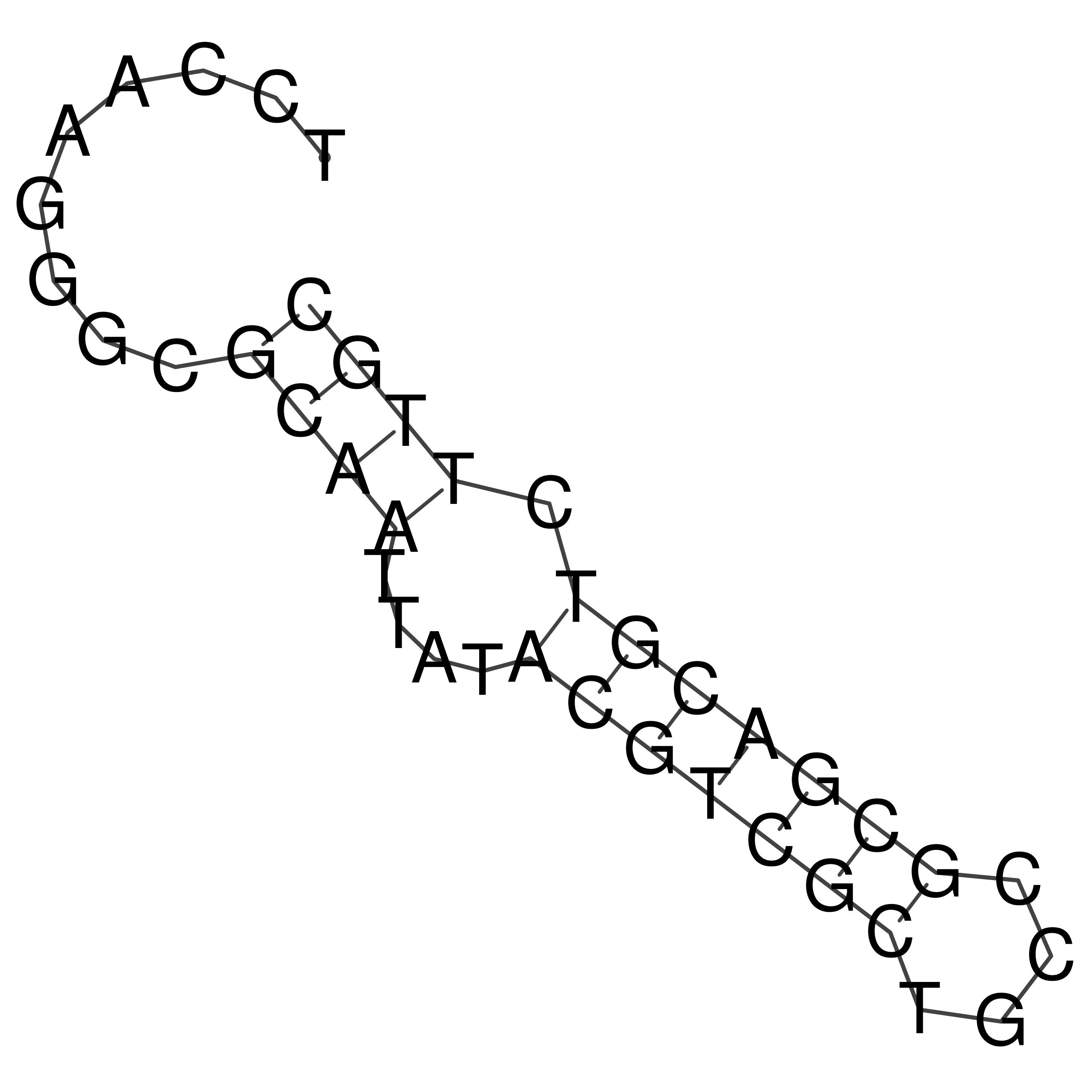
Relaxase
| ID | 4593 | GenBank | WP_280736468 |
| Name | traA_PYH37_RS32140_CIP 109850|unnamed |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 122318.05 Da Isoelectric Point: 9.6543
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYRAKQGLLHEEFVMPADSPEWLRSMIADR
SVAGASEAFWNEVEDFEKRADAQLAKDVTIALPIELSAEQNIALIRDFVELHVTAKGMVADWVYHDAPGN
PHVHLMTTLRPLTDDGFGGKKIAALGPDGNPLRNDAGKIVYELWAGGLDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKSATDEQAASLERLELQEEKRAENARRIQRRPGIVLGV
ITREKSVFDERDVAKVLYRYIDDPGIFQSLMARILQSPETLRLEREQVVFATGVRTPAKYTTRGLILLEG
EMANRAIWLSTRSSHGVRKEVLEATFTRHSRLSTEQKTAIEHMAGAERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAESLEKEAGIVSRTLSSWELRWSEGRNQLDDKAVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLIGDPEQLQPIEAGAAFRAIADRLGYAELETIYRQREQWMRDASLDLARGNVGKAVDAYR
ANGKMMGSELKAQAVENLIADWDRDYDPTKTTLILAHLRRDVRMLNEMARAKLVERGIVGEGFVFRTEDG
NRRFAPGDQIVFLKNEGSLGVKNGMLGKVVDASKNRIVAEIGEGEHRRQVTVEQRFYNNLDHGYATTIHK
SQGATVDRLKVLASLSLDRHLTYVAMTRHREDLGVYYGARSFAKSGGLIPILSRENSKETTLDYERATFY
RQALHFAEARGLHLVNVARTIARDRLEWTVRQKQKLADLAHRLVAIGARLGLAVGAKPTRSQSIKETRPM
VAGIATFPKSIEQAVADKLEADPGLKKQWEDVSTRFRLVYAQPEAAFKAVNVDAVLKDDTVARSTLATIA
SQPESYGALKGKTGLLTSRADKQDRERAERNAPALAQSLSDYMRQRSQAEQRYQAEELAARRKVAIDIPA
LSSSARQTLERVRDAIDRNDLPAALEFALADREVKAELEGFAKTVTERFGERTFLPLAAKDTNGETFRTI
TAGMKPGQKAEVQSAWNTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4688 | GenBank | WP_280736465 |
| Name | traG_PYH37_RS29170_CIP 109850|unnamed |
UniProt ID | _ |
| Length | 660 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 660 a.a. Molecular weight: 71251.27 Da Isoelectric Point: 9.0510
MTANRLALVAVPVVLMILAVIGMTGIEQWLSAFGKTDAARLTLGRVGIAAPYVTAAAIGLIFLFASAGST
SIKFAGWGALAGSAATMLVAAIREATRLAGFAGQVPADKSIASYLDPATIIGAASALIAGCFALRVALIG
NVAFARAEPKRIRGQRALHGQADWMKMQDAAKLFPDTGGIVVGERYRVDKDSTAARPFRADDPETWATGG
NSPLLCFDGSFGSSHGIVFAGSGGFKTTSATIPTALKWGGGLVVLDPSNEVAPMVYEHRKNAARFIRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIMSDSGGPRGVRDDFFRASALQLLTALIADVCLSGHTDT
ENQTLRQVRVNLSEPEPKLRERLQSIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFSTDALAAGDTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGELKGRALFLLDEVARLGY
MRILETARDAGRRYGISLTMIYQSIGQMRETYGGRDAASKWFESASWISFASINDPDTADYISRRCGMTT
VEIDQVSRTFQAKGSSRTRSKQLAARPLIQAHEVLRMRADEQIVFTAGNPPLRCGRAIWFRREDMKACVG
TNRFHRIGKPSDARVIEPARCATSKADPGK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4689 | GenBank | WP_280736383 |
| Name | virD4_PYH37_RS29685_CIP 109850|unnamed |
UniProt ID | _ |
| Length | 569 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 569 a.a. Molecular weight: 63582.95 Da Isoelectric Point: 8.8049
MISTETRPLSIAVSIVCSLAAGFCGASLYSTFRYGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGTILLLQQMIPGRNRQHHGTARWARVDEMRRAGYLQRYNRISGPVFGKTSARFWSDYYLTNSEQP
HSLVVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKARGDSIFKLAPLDPERRTNCYNPL
LDIIAMSPERQFTEARRLAANLITAKGQGAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
RLFARLAEETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPLVKAATSRSDFSIYDLRRRRTC
VYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYDEAGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYSQGRMFDRNI
QNSDQGAPLLRPEQVRLLSDKYEIVLIKGQPPLQLQKVRYYSDRALKHIFESQTGDLPEPAPLMIEEGKL
GVNSSFNDC
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4690 | GenBank | WP_280736473 |
| Name | t4cp2_PYH37_RS32080_CIP 109850|unnamed |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91573.23 Da Isoelectric Point: 5.6675
MVALKRFRAPGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
SGWMIQVEAVRIPTFDYASEDRCHFSDAVTHAIDAERRAHFARERGHFESKHAVILTYRPVESKKTALSK
YIYSDEESRKKTYADTVLFIFKNAIREIEQYLANTLSIRRMETREVAERGGERVARYDELLQFVRFCITG
ENHPIRLPDAPMYLDWIATAELEHGLTPKVENQFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVIIL
FDSDREALQEKAEAIRRLIQAEGFGGRIETLNATDAYLGSLPGNWYSNIREPLINTSNLADLVPLNSVWS
GNPSAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTMIFGPTGSGKSTLLALIAAQFRRYNQAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDAHDAGPPLAFCPLSDLGSDSDRAWATEWIEMLIALQGITITPDHRN
AISRQVTLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEDDGLALGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNSAAREPGTREFYERIGFNERQIEIVSNAIPKREYYVVTPEGRRLFDMALGP
VALSFVGASGKEDLKRIRTLQSEHGRDWPVHWLQMRGVHDAASLLEVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 101101..110524
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PYH37_RS29600 (PYH37_005920) | 97908..98255 | + | 348 | WP_280736394 | hypothetical protein | - |
| PYH37_RS29605 (PYH37_005921) | 98528..98791 | + | 264 | Protein_87 | transposase | - |
| PYH37_RS29610 (PYH37_005922) | 99048..99650 | + | 603 | WP_280736393 | hypothetical protein | - |
| PYH37_RS29615 (PYH37_005923) | 99792..100031 | - | 240 | WP_280736392 | helix-turn-helix transcriptional regulator | - |
| PYH37_RS29620 (PYH37_005924) | 101101..101808 | + | 708 | WP_280736391 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| PYH37_RS29625 (PYH37_005925) | 101828..102190 | + | 363 | WP_280663464 | pilin major subunit VirB2 | virB2 |
| PYH37_RS29630 (PYH37_005926) | 102190..102516 | + | 327 | WP_280663465 | type IV secretion system protein VirB3 | virB3 |
| PYH37_RS29635 (PYH37_005927) | 102516..104885 | + | 2370 | WP_280736390 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| PYH37_RS29640 (PYH37_005928) | 104899..105579 | + | 681 | WP_280736389 | pilin minor subunit VirB5 | - |
| PYH37_RS29645 (PYH37_005929) | 105658..106545 | + | 888 | WP_280736388 | type IV secretion system protein | virB6 |
| PYH37_RS29650 (PYH37_005930) | 106579..106746 | + | 168 | WP_194732549 | type IV secretion system lipoprotein VirB7 | - |
| PYH37_RS29655 (PYH37_005931) | 106733..107446 | + | 714 | WP_280663469 | type IV secretion system protein VirB8 | virB8 |
| PYH37_RS29660 (PYH37_005932) | 107443..108324 | + | 882 | WP_280736387 | P-type conjugative transfer protein VirB9 | virB9 |
| PYH37_RS29665 (PYH37_005933) | 108321..109445 | + | 1125 | WP_280736386 | type IV secretion system protein VirB10 | virB10 |
| PYH37_RS29670 (PYH37_005934) | 109484..110524 | + | 1041 | WP_280736385 | P-type DNA transfer ATPase VirB11 | virB11 |
| PYH37_RS29675 (PYH37_005935) | 111595..112299 | + | 705 | WP_280736604 | response regulator | - |
| PYH37_RS29680 (PYH37_005936) | 112408..112602 | - | 195 | WP_280736384 | hypothetical protein | - |
| PYH37_RS29685 (PYH37_005937) | 112739..114448 | - | 1710 | WP_280736383 | type IV secretion system ATPase VirD4 | - |
Host bacterium
| ID | 7389 | GenBank | NZ_CP120369 |
| Plasmid name | CIP 109850|unnamed | Incompatibility group | - |
| Plasmid size | 629445 bp | Coordinate of oriT [Strand] | 628761..628800 [-] |
| Host baterium | Sinorhizobium numidicum strain CIP 109850 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | linJ, dxnH |
| Symbiosis gene | fixX, fixC, fixB, fixA, nifW, nifS, nifH, nifD, nifE, nifX, mLTONO_5203, nodD, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, nifN, nifB, nifZ, fixU, a9174_31165, nodM, fixS, fixG, fixO, fixN |
| Anti-CRISPR | AcrIIA9, AcrIIC2 |