Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106951 |
| Name | oriT_LMG 27395|unnamed |
| Organism | Sinorhizobium numidicum strain LMG 27395 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP120372 (136214..136253 [+], 40 nt) |
| oriT length | 40 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 40 nt
TCCAAGGGCGCAATTATACGTCGCTGCCGCGACGTCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file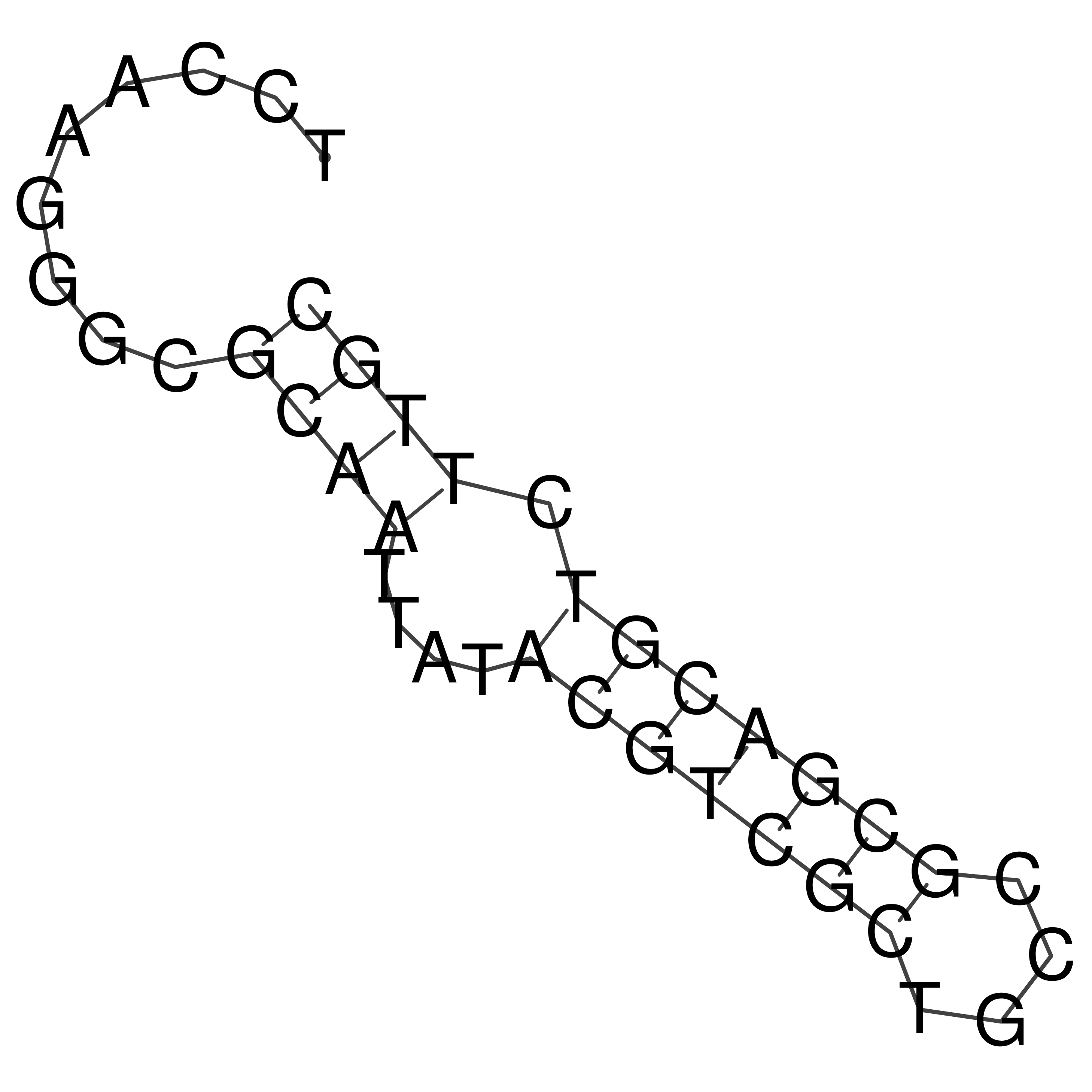
Relaxase
| ID | 4592 | GenBank | WP_280736468 |
| Name | traA_PYH38_RS29785_LMG 27395|unnamed |
UniProt ID | _ |
| Length | 1102 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1102 a.a. Molecular weight: 122318.05 Da Isoelectric Point: 9.6543
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEFEREARTIDYRAKQGLLHEEFVMPADSPEWLRSMIADR
SVAGASEAFWNEVEDFEKRADAQLAKDVTIALPIELSAEQNIALIRDFVELHVTAKGMVADWVYHDAPGN
PHVHLMTTLRPLTDDGFGGKKIAALGPDGNPLRNDAGKIVYELWAGGLDDFNAFRDGWFACQNRHLALAG
LDIRIDGRSFEKQGIELTPTIHLGVGTKAIERKSATDEQAASLERLELQEEKRAENARRIQRRPGIVLGV
ITREKSVFDERDVAKVLYRYIDDPGIFQSLMARILQSPETLRLEREQVVFATGVRTPAKYTTRGLILLEG
EMANRAIWLSTRSSHGVRKEVLEATFTRHSRLSTEQKTAIEHMAGAERIAAVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKAAESLEKEAGIVSRTLSSWELRWSEGRNQLDDKAVFVLDEAGMVSSRQMALFVE
AATKAGAKLVLIGDPEQLQPIEAGAAFRAIADRLGYAELETIYRQREQWMRDASLDLARGNVGKAVDAYR
ANGKMMGSELKAQAVENLIADWDRDYDPTKTTLILAHLRRDVRMLNEMARAKLVERGIVGEGFVFRTEDG
NRRFAPGDQIVFLKNEGSLGVKNGMLGKVVDASKNRIVAEIGEGEHRRQVTVEQRFYNNLDHGYATTIHK
SQGATVDRLKVLASLSLDRHLTYVAMTRHREDLGVYYGARSFAKSGGLIPILSRENSKETTLDYERATFY
RQALHFAEARGLHLVNVARTIARDRLEWTVRQKQKLADLAHRLVAIGARLGLAVGAKPTRSQSIKETRPM
VAGIATFPKSIEQAVADKLEADPGLKKQWEDVSTRFRLVYAQPEAAFKAVNVDAVLKDDTVARSTLATIA
SQPESYGALKGKTGLLTSRADKQDRERAERNAPALAQSLSDYMRQRSQAEQRYQAEELAARRKVAIDIPA
LSSSARQTLERVRDAIDRNDLPAALEFALADREVKAELEGFAKTVTERFGERTFLPLAAKDTNGETFRTI
TAGMKPGQKAEVQSAWNTMRTVQQLSAHERTTEALKQAETLRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4685 | GenBank | WP_280736383 |
| Name | virD4_PYH38_RS29250_LMG 27395|unnamed |
UniProt ID | _ |
| Length | 569 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 569 a.a. Molecular weight: 63582.95 Da Isoelectric Point: 8.8049
MISTETRPLSIAVSIVCSLAAGFCGASLYSTFRYGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGTILLLQQMIPGRNRQHHGTARWARVDEMRRAGYLQRYNRISGPVFGKTSARFWSDYYLTNSEQP
HSLVVAPTRAGKGVGIVIPTLLTFKGSVIALDVKGELFELTSRARKARGDSIFKLAPLDPERRTNCYNPL
LDIIAMSPERQFTEARRLAANLITAKGQGAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
RLFARLAEETRNKEAQRIFDDMAGNDTKILTSYTSVLGDGGLNLWADPLVKAATSRSDFSIYDLRRRRTC
VYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYDEAGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYSQGRMFDRNI
QNSDQGAPLLRPEQVRLLSDKYEIVLIKGQPPLQLQKVRYYSDRALKHIFESQTGDLPEPAPLMIEEGKL
GVNSSFNDC
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4686 | GenBank | WP_280736465 |
| Name | traG_PYH38_RS29770_LMG 27395|unnamed |
UniProt ID | _ |
| Length | 660 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 660 a.a. Molecular weight: 71251.27 Da Isoelectric Point: 9.0510
MTANRLALVAVPVVLMILAVIGMTGIEQWLSAFGKTDAARLTLGRVGIAAPYVTAAAIGLIFLFASAGST
SIKFAGWGALAGSAATMLVAAIREATRLAGFAGQVPADKSIASYLDPATIIGAASALIAGCFALRVALIG
NVAFARAEPKRIRGQRALHGQADWMKMQDAAKLFPDTGGIVVGERYRVDKDSTAARPFRADDPETWATGG
NSPLLCFDGSFGSSHGIVFAGSGGFKTTSATIPTALKWGGGLVVLDPSNEVAPMVYEHRKNAARFIRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIMSDSGGPRGVRDDFFRASALQLLTALIADVCLSGHTDT
ENQTLRQVRVNLSEPEPKLRERLQSIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFSTDALAAGDTDVFINIDLKTLETHSGLARVIIGSFLNAIYNRDGELKGRALFLLDEVARLGY
MRILETARDAGRRYGISLTMIYQSIGQMRETYGGRDAASKWFESASWISFASINDPDTADYISRRCGMTT
VEIDQVSRTFQAKGSSRTRSKQLAARPLIQAHEVLRMRADEQIVFTAGNPPLRCGRAIWFRREDMKACVG
TNRFHRIGKPSDARVIEPARCATSKADPGK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4687 | GenBank | WP_280736473 |
| Name | t4cp2_PYH38_RS29845_LMG 27395|unnamed |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91573.23 Da Isoelectric Point: 5.6675
MVALKRFRAPGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAILSRLG
SGWMIQVEAVRIPTFDYASEDRCHFSDAVTHAIDAERRAHFARERGHFESKHAVILTYRPVESKKTALSK
YIYSDEESRKKTYADTVLFIFKNAIREIEQYLANTLSIRRMETREVAERGGERVARYDELLQFVRFCITG
ENHPIRLPDAPMYLDWIATAELEHGLTPKVENQFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVIIL
FDSDREALQEKAEAIRRLIQAEGFGGRIETLNATDAYLGSLPGNWYSNIREPLINTSNLADLVPLNSVWS
GNPSAPCPFYPPNSPPLMQVASGSTPFRLNLHVDDVGHTMIFGPTGSGKSTLLALIAAQFRRYNQAQIFA
FDKGSSLLPLTLAAGGDHYEIGGDAHDAGPPLAFCPLSDLGSDSDRAWATEWIEMLIALQGITITPDHRN
AISRQVTLMASAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEDDGLALGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNSAAREPGTREFYERIGFNERQIEIVSNAIPKREYYVVTPEGRRLFDMALGP
VALSFVGASGKEDLKRIRTLQSEHGRDWPVHWLQMRGVHDAASLLEVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 25045..34468
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PYH38_RS29250 (PYH38_005849) | 21121..22830 | + | 1710 | WP_280736383 | type IV secretion system ATPase VirD4 | - |
| PYH38_RS29255 (PYH38_005850) | 22967..23161 | + | 195 | WP_280736384 | hypothetical protein | - |
| PYH38_RS29260 (PYH38_005851) | 23270..23974 | - | 705 | WP_280736604 | response regulator | - |
| PYH38_RS29265 (PYH38_005852) | 25045..26085 | - | 1041 | WP_280736385 | P-type DNA transfer ATPase VirB11 | virB11 |
| PYH38_RS29270 (PYH38_005853) | 26124..27248 | - | 1125 | WP_280736386 | type IV secretion system protein VirB10 | virB10 |
| PYH38_RS29275 (PYH38_005854) | 27245..28126 | - | 882 | WP_280736387 | P-type conjugative transfer protein VirB9 | virB9 |
| PYH38_RS29280 (PYH38_005855) | 28123..28836 | - | 714 | WP_280663469 | type IV secretion system protein VirB8 | virB8 |
| PYH38_RS29285 (PYH38_005856) | 28823..28990 | - | 168 | WP_194732549 | type IV secretion system lipoprotein VirB7 | - |
| PYH38_RS29290 (PYH38_005857) | 29024..29911 | - | 888 | WP_280736388 | type IV secretion system protein | virB6 |
| PYH38_RS29295 (PYH38_005858) | 29990..30670 | - | 681 | WP_280736389 | pilin minor subunit VirB5 | - |
| PYH38_RS29300 (PYH38_005859) | 30684..33053 | - | 2370 | WP_280736390 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| PYH38_RS29305 (PYH38_005860) | 33053..33379 | - | 327 | WP_280663465 | type IV secretion system protein VirB3 | virB3 |
| PYH38_RS29310 (PYH38_005861) | 33379..33741 | - | 363 | WP_280663464 | pilin major subunit VirB2 | virB2 |
| PYH38_RS29315 (PYH38_005862) | 33761..34468 | - | 708 | WP_280736391 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| PYH38_RS29320 (PYH38_005863) | 35538..35777 | + | 240 | WP_280736392 | helix-turn-helix transcriptional regulator | - |
| PYH38_RS29325 (PYH38_005864) | 35919..36521 | - | 603 | WP_280736393 | hypothetical protein | - |
| PYH38_RS29330 (PYH38_005865) | 36778..37041 | - | 264 | Protein_34 | transposase | - |
| PYH38_RS29335 (PYH38_005866) | 37314..37661 | - | 348 | WP_280736394 | hypothetical protein | - |
Host bacterium
| ID | 7388 | GenBank | NZ_CP120372 |
| Plasmid name | LMG 27395|unnamed | Incompatibility group | - |
| Plasmid size | 629446 bp | Coordinate of oriT [Strand] | 136214..136253 [+] |
| Host baterium | Sinorhizobium numidicum strain LMG 27395 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | dxnH, linJ |
| Symbiosis gene | nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, mLTONO_5203, nifX, nifE, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, fixN, fixO, fixG, fixS, nodM, a9174_31165, fixU, nifZ, nifB |
| Anti-CRISPR | AcrIIC2, AcrIIA9 |