Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106948 |
| Name | oriT_psymB |
| Organism | Sinorhizobium meliloti strain M270 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021814 (918092..918120 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_psymB
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file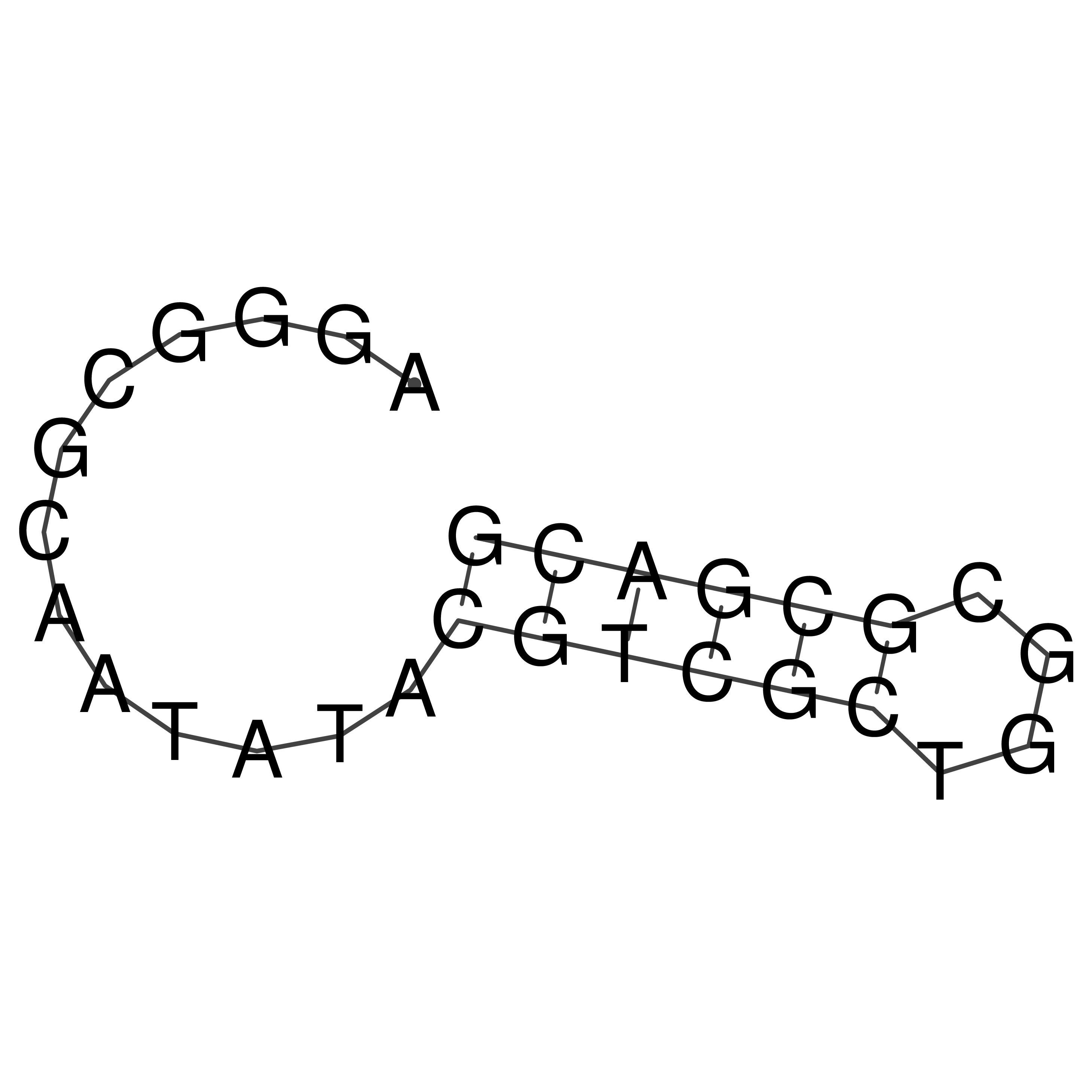
Relaxase
| ID | 4588 | GenBank | WP_088198608 |
| Name | traA_CDO26_RS30085_psymB |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169908.58 Da Isoelectric Point: 9.5024
>WP_088198608.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMAALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADFARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDELVLLKPQQVDAETGKAKQPAVFTTREMLRIE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDALQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARSK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRQVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPDWGRRPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRIQSSQKIVSERRQRAINQSRGIRM
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMAALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADFARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDELVLLKPQQVDAETGKAKQPAVFTTREMLRIE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDALQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARSK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRQVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPDWGRRPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRIQSSQKIVSERRQRAINQSRGIRM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4679 | GenBank | WP_094183236 |
| Name | tcpA_CDO26_RS37485_psymB |
UniProt ID | _ |
| Length | 947 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 947 a.a. Molecular weight: 101760.56 Da Isoelectric Point: 4.6250
>WP_094183236.1 FtsK/SpoIIIE domain-containing protein [Sinorhizobium meliloti]
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
RAARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEE
SRIEADEAAAEASDAETVMDVVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPA
ITESVAVAEEGPATSETTALPVTDEVPVVPHALPVANLAPESAAVEEDVPHQADARLAYLSDFAFFEFMP
LEVAAVPPTVTEPVKEAARIPAPIAAAPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEP
VTAAPQDIAAEPAAAEAAALPAHQFVETPAPALAEPAIVPASEPAPQAPVTRAAITMPAVIQRSSPSLPP
IGAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYE
FEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLA
LCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGI
PHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTG
EPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIK
ANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRP
EYLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLV
ERMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
RAARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEE
SRIEADEAAAEASDAETVMDVVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPA
ITESVAVAEEGPATSETTALPVTDEVPVVPHALPVANLAPESAAVEEDVPHQADARLAYLSDFAFFEFMP
LEVAAVPPTVTEPVKEAARIPAPIAAAPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEP
VTAAPQDIAAEPAAAEAAALPAHQFVETPAPALAEPAIVPASEPAPQAPVTRAAITMPAVIQRSSPSLPP
IGAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYE
FEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLA
LCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGI
PHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTG
EPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIK
ANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRP
EYLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLV
ERMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 7385 | GenBank | NZ_CP021814 |
| Plasmid name | psymB | Incompatibility group | - |
| Plasmid size | 1694441 bp | Coordinate of oriT [Strand] | 918092..918120 [-] |
| Host baterium | Sinorhizobium meliloti strain M270 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |