Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106946 |
| Name | oriT_psymA |
| Organism | Sinorhizobium meliloti strain M270 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021813 (995932..995960 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_psymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file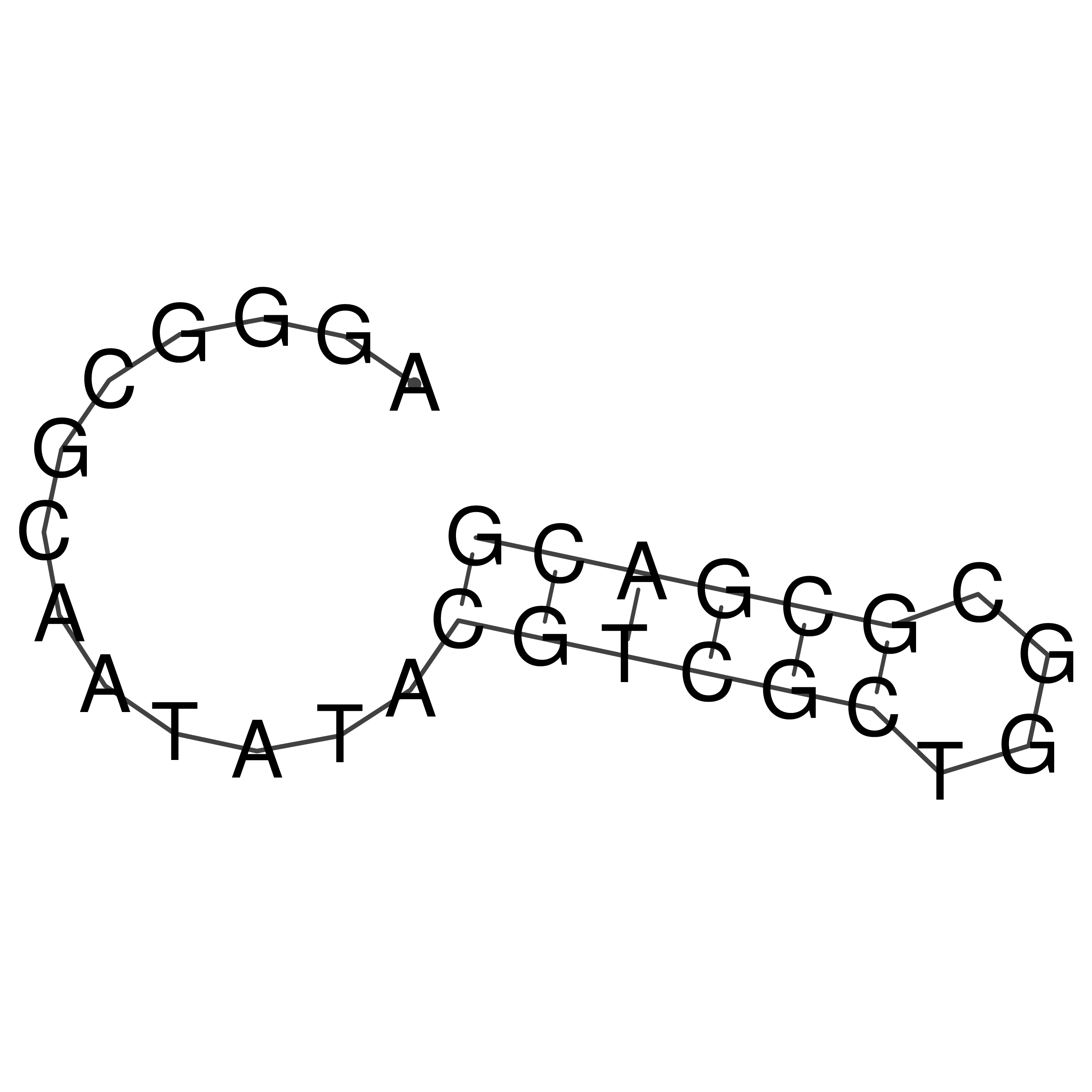
Relaxase
| ID | 4587 | GenBank | WP_088198164 |
| Name | traA_CDO26_RS23385_psymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169919.33 Da Isoelectric Point: 9.0340
>WP_088198164.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSSVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKSQQIDAETGKAKQPAVFTTREMLRLE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDGWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
TSLRQVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTQLSVRLDSGRDVIISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPDWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVADDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERIAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAVGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRVQSSQEIISERQRRVIDRARGVTR
MAIMFVRAQVIGRGAGRSSVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKSQQIDAETGKAKQPAVFTTREMLRLE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDGWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
TSLRQVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTQLSVRLDSGRDVIISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPDWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVADDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERIAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAVGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRVQSSQEIISERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4677 | GenBank | WP_088198162 |
| Name | traG_CDO26_RS23370_psymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70281.21 Da Isoelectric Point: 9.3594
>WP_088198162.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLWVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWVL
PLHRRRPVAIASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFDPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRLESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNTGWDMNNSASRKSESVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLWVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWVL
PLHRRRPVAIASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFDPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRLESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNTGWDMNNSASRKSESVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1096086..1105665
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CDO26_RS23845 (CDO26_23845) | 1092409..1092696 | - | 288 | WP_013845449 | chaperone NapD | - |
| CDO26_RS23850 (CDO26_23850) | 1092689..1093189 | - | 501 | WP_088198220 | ferredoxin-type protein NapF | - |
| CDO26_RS23855 (CDO26_23855) | 1093189..1093374 | - | 186 | WP_088198221 | periplasmic nitrate reductase, NapE protein | - |
| CDO26_RS23860 (CDO26_23860) | 1093901..1094512 | + | 612 | WP_028003331 | TetR/AcrR family transcriptional regulator | - |
| CDO26_RS23865 (CDO26_23865) | 1094549..1095775 | + | 1227 | WP_013845453 | MFS transporter | - |
| CDO26_RS23870 (CDO26_23870) | 1096086..1097075 | - | 990 | WP_088198222 | P-type DNA transfer ATPase VirB11 | virB11 |
| CDO26_RS23875 (CDO26_23875) | 1097086..1098258 | - | 1173 | WP_013845455 | type IV secretion system protein VirB10 | virB10 |
| CDO26_RS23880 (CDO26_23880) | 1098268..1099122 | - | 855 | WP_014531548 | P-type conjugative transfer protein VirB9 | virB9 |
| CDO26_RS23885 (CDO26_23885) | 1099122..1099793 | - | 672 | WP_013845457 | virB8 family protein | virB8 |
| CDO26_RS23890 (CDO26_23890) | 1099798..1100112 | - | 315 | WP_013845458 | hypothetical protein | - |
| CDO26_RS23895 (CDO26_23895) | 1100109..1101044 | - | 936 | WP_167693537 | type IV secretion system protein | virB6 |
| CDO26_RS23900 (CDO26_23900) | 1101050..1101283 | - | 234 | WP_088198224 | EexN family lipoprotein | - |
| CDO26_RS23905 (CDO26_23905) | 1101280..1101981 | - | 702 | WP_088198225 | P-type DNA transfer protein VirB5 | virB5 |
| CDO26_RS23910 (CDO26_23910) | 1101981..1104377 | - | 2397 | WP_207953757 | VirB4 family type IV secretion system protein | virb4 |
| CDO26_RS23915 (CDO26_23915) | 1104352..1104693 | - | 342 | WP_017264162 | type IV secretion system protein VirB3 | virB3 |
| CDO26_RS23920 (CDO26_23920) | 1104698..1104997 | - | 300 | WP_003526741 | TrbC/VirB2 family protein | virB2 |
| CDO26_RS23925 (CDO26_23925) | 1104994..1105665 | - | 672 | WP_088198227 | lytic transglycosylase domain-containing protein | virB1 |
| CDO26_RS23930 (CDO26_23930) | 1105669..1106187 | - | 519 | WP_017277528 | hypothetical protein | - |
| CDO26_RS23935 (CDO26_23935) | 1106284..1106652 | + | 369 | WP_010967698 | transcriptional regulator | - |
| CDO26_RS23940 (CDO26_23940) | 1106913..1107164 | - | 252 | WP_088198228 | hypothetical protein | - |
| CDO26_RS23945 (CDO26_23945) | 1107238..1107885 | - | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| CDO26_RS23950 (CDO26_23950) | 1107935..1108783 | - | 849 | WP_088198229 | alpha/beta fold hydrolase | - |
Host bacterium
| ID | 7383 | GenBank | NZ_CP021813 |
| Plasmid name | psymA | Incompatibility group | - |
| Plasmid size | 1454983 bp | Coordinate of oriT [Strand] | 995932..995960 [+] |
| Host baterium | Sinorhizobium meliloti strain M270 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | ncrA |
| Degradation gene | - |
| Symbiosis gene | nodD, fixN, fixO, fixG, nifB, fixX, fixC, fixB, fixA, nifX, nifE, nifK, nifD, nifH, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, nifN, nodM |
| Anti-CRISPR | AcrIIA9 |