Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106913 |
| Name | oriT_p1_045523 |
| Organism | Enterobacter kobei strain WCHEK045523 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP032893 (135986..136088 [-], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 89..94, 96..101 (AAATAA..TTATTT) 28..33, 40..45 (GCAAAA..TTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_p1_045523
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTGCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTGCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file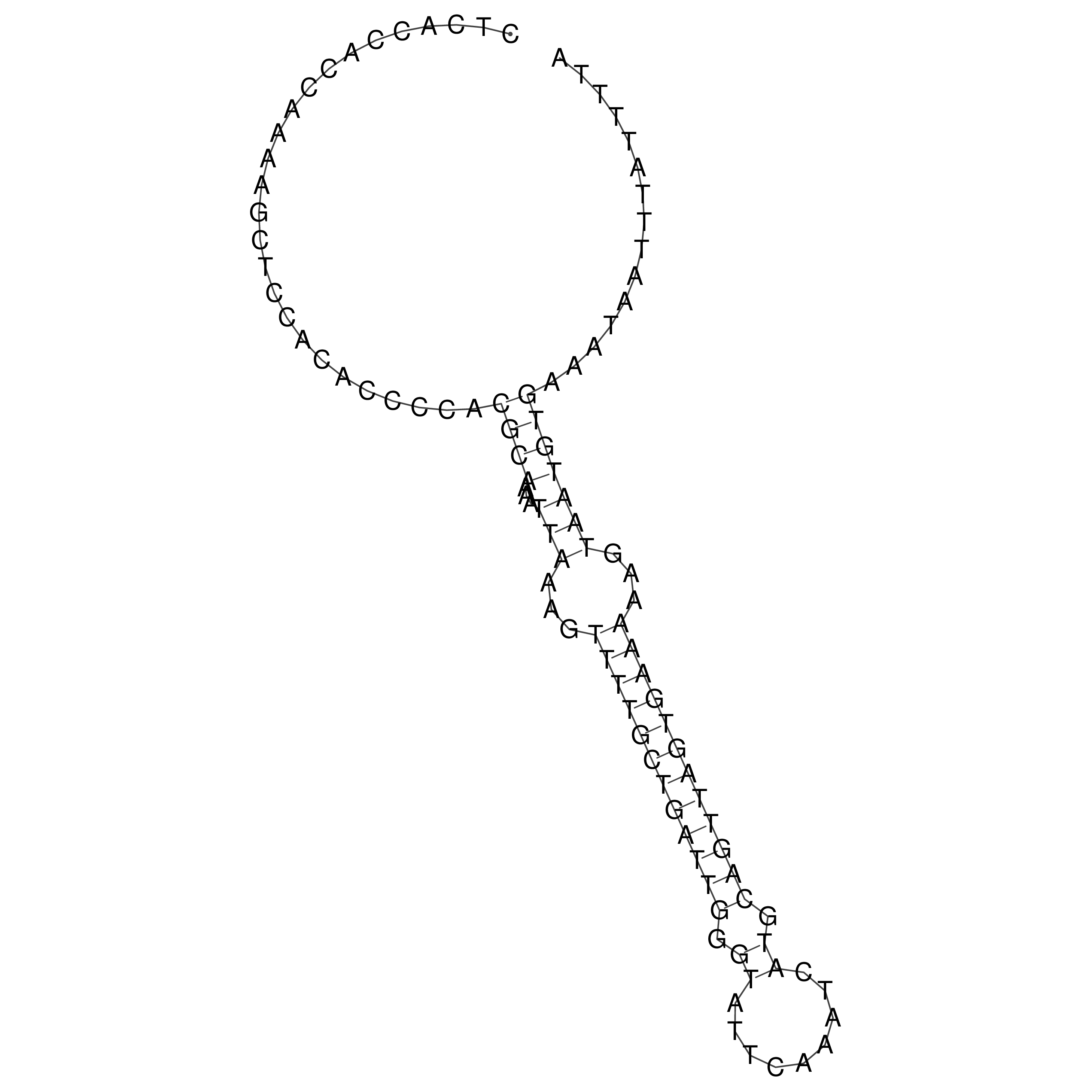
Relaxase
| ID | 4570 | GenBank | WP_023307204 |
| Name | traI_D9T11_RS00590_p1_045523 |
UniProt ID | A0A1Q1PS85 |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189264.44 Da Isoelectric Point: 6.1121
>WP_023307204.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4655 | GenBank | WP_008324910 |
| Name | traD_D9T11_RS00595_p1_045523 |
UniProt ID | _ |
| Length | 730 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 730 a.a. Molecular weight: 81859.42 Da Isoelectric Point: 5.1609
>WP_008324910.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQAEQDE
LSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQAEQDE
LSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4656 | GenBank | WP_008324134 |
| Name | traC_D9T11_RS00670_p1_045523 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98841.51 Da Isoelectric Point: 5.7676
>WP_008324134.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacterales]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 111520..136636
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| D9T11_RS00595 (D9T11_00595) | 111520..113712 | - | 2193 | WP_008324910 | type IV conjugative transfer system coupling protein TraD | virb4 |
| D9T11_RS00600 (D9T11_00600) | 113846..114520 | - | 675 | WP_008324909 | tetratricopeptide repeat protein | - |
| D9T11_RS00605 (D9T11_00605) | 114595..115026 | - | 432 | WP_008324118 | entry exclusion protein | - |
| D9T11_RS00610 (D9T11_00610) | 115048..117870 | - | 2823 | WP_008324119 | conjugal transfer mating-pair stabilization protein TraG | traG |
| D9T11_RS00615 (D9T11_00615) | 117870..119240 | - | 1371 | WP_016247527 | conjugal transfer pilus assembly protein TraH | traH |
| D9T11_RS00620 (D9T11_00620) | 119227..119655 | - | 429 | WP_008324122 | hypothetical protein | - |
| D9T11_RS00625 (D9T11_00625) | 119648..120202 | - | 555 | WP_008324125 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| D9T11_RS00630 (D9T11_00630) | 120192..120440 | - | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| D9T11_RS00635 (D9T11_00635) | 120457..121206 | - | 750 | WP_008324127 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| D9T11_RS00640 (D9T11_00640) | 121237..121425 | - | 189 | WP_008324128 | conjugal transfer protein TrbE | - |
| D9T11_RS00645 (D9T11_00645) | 121457..123310 | - | 1854 | WP_022652255 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| D9T11_RS00650 (D9T11_00650) | 123307..123936 | - | 630 | WP_008324130 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| D9T11_RS00655 (D9T11_00655) | 123953..124939 | - | 987 | WP_032160671 | conjugal transfer pilus assembly protein TraU | traU |
| D9T11_RS00660 (D9T11_00660) | 124942..125589 | - | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| D9T11_RS00665 (D9T11_00665) | 125589..125936 | - | 348 | WP_008324133 | type-F conjugative transfer system protein TrbI | - |
| D9T11_RS00670 (D9T11_00670) | 125933..128563 | - | 2631 | WP_008324134 | type IV secretion system protein TraC | virb4 |
| D9T11_RS00675 (D9T11_00675) | 128556..129122 | - | 567 | WP_008324135 | conjugal transfer pilus-stabilizing protein TraP | - |
| D9T11_RS00680 (D9T11_00680) | 129112..129513 | - | 402 | WP_008324137 | hypothetical protein | - |
| D9T11_RS00685 (D9T11_00685) | 129497..129682 | - | 186 | WP_008324139 | hypothetical protein | - |
| D9T11_RS00690 (D9T11_00690) | 129686..129910 | - | 225 | WP_008324141 | TraR/DksA C4-type zinc finger protein | - |
| D9T11_RS00695 (D9T11_00695) | 130030..130569 | - | 540 | WP_008324154 | type IV conjugative transfer system lipoprotein TraV | traV |
| D9T11_RS00700 (D9T11_00700) | 130591..131985 | - | 1395 | WP_008324156 | F-type conjugal transfer pilus assembly protein TraB | traB |
| D9T11_RS00705 (D9T11_00705) | 131972..132715 | - | 744 | WP_008324157 | type-F conjugative transfer system secretin TraK | traK |
| D9T11_RS00710 (D9T11_00710) | 132702..133268 | - | 567 | WP_008324158 | type IV conjugative transfer system protein TraE | traE |
| D9T11_RS00715 (D9T11_00715) | 133283..133588 | - | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| D9T11_RS00720 (D9T11_00720) | 133739..134089 | - | 351 | WP_008324162 | type IV conjugative transfer system pilin TraA | - |
| D9T11_RS00725 (D9T11_00725) | 134147..134371 | - | 225 | WP_008324165 | TraY domain-containing protein | - |
| D9T11_RS00730 (D9T11_00730) | 134458..135147 | - | 690 | WP_008324166 | hypothetical protein | - |
| D9T11_RS00735 (D9T11_00735) | 135336..135728 | - | 393 | WP_008324167 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| D9T11_RS00740 (D9T11_00740) | 136160..136636 | + | 477 | WP_008324168 | transglycosylase SLT domain-containing protein | virB1 |
| D9T11_RS00745 (D9T11_00745) | 136683..137213 | - | 531 | WP_008324170 | antirestriction protein | - |
| D9T11_RS00750 (D9T11_00750) | 137840..138670 | - | 831 | WP_008324171 | N-6 DNA methylase | - |
| D9T11_RS00755 (D9T11_00755) | 138713..138976 | - | 264 | WP_008324174 | hypothetical protein | - |
| D9T11_RS00760 (D9T11_00760) | 139062..139406 | - | 345 | WP_008324177 | hypothetical protein | - |
| D9T11_RS00765 (D9T11_00765) | 139453..139740 | - | 288 | WP_008324178 | hypothetical protein | - |
| D9T11_RS00770 (D9T11_00770) | 139794..140168 | - | 375 | WP_008324180 | hypothetical protein | - |
| D9T11_RS00775 (D9T11_00775) | 140795..141154 | - | 360 | WP_008324181 | hypothetical protein | - |
| D9T11_RS00780 (D9T11_00780) | 141216..141539 | - | 324 | WP_008324183 | hypothetical protein | - |
Host bacterium
| ID | 7350 | GenBank | NZ_CP032893 |
| Plasmid name | p1_045523 | Incompatibility group | IncFIB |
| Plasmid size | 156442 bp | Coordinate of oriT [Strand] | 135986..136088 [-] |
| Host baterium | Enterobacter kobei strain WCHEK045523 |
Cargo genes
| Drug resistance gene | mcr-9, aadA2, qacE, sul1 |
| Virulence gene | - |
| Metal resistance gene | rcnR/yohL, ncrC, pcoE, pcoS, silP, silA, silB, silF, silC, silR, silS, silE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |