Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106902 |
| Name | oriT_Lep1A1|p5 |
| Organism | Caballeronia grimmiae strain Lep1A1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP084278 (41960..42058 [-], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_Lep1A1|p5
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file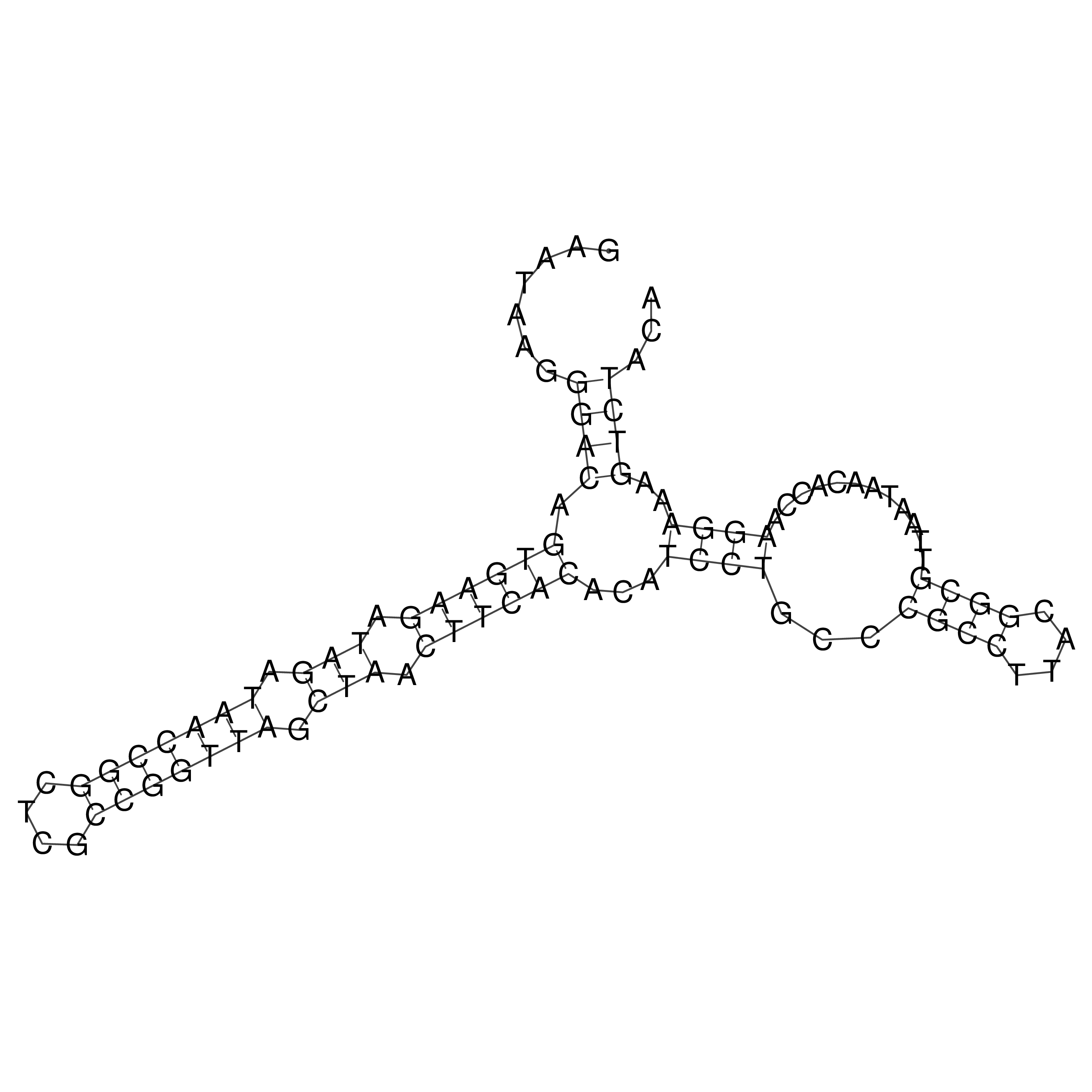
Relaxase
| ID | 4563 | GenBank | WP_011114057 |
| Name | Relaxase_LDZ21_RS31050_Lep1A1|p5 |
UniProt ID | A0A4P7LSA6 |
| Length | 746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 82038.71 Da Isoelectric Point: 10.7900
>WP_011114057.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Bacteria]
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
LQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
LQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A4P7LSA6 |
T4CP
| ID | 4649 | GenBank | WP_011114047 |
| Name | t4cp2_LDZ21_RS30820_Lep1A1|p5 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93980.15 Da Isoelectric Point: 6.3901
>WP_011114047.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Bacteria]
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4650 | GenBank | WP_000694765 |
| Name | t4cp2_LDZ21_RS31055_Lep1A1|p5 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69897.09 Da Isoelectric Point: 8.2993
>WP_000694765.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Bacteria]
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 7339 | GenBank | NZ_CP084278 |
| Plasmid name | Lep1A1|p5 | Incompatibility group | IncP1 |
| Plasmid size | 65095 bp | Coordinate of oriT [Strand] | 41960..42058 [-] |
| Host baterium | Caballeronia grimmiae strain Lep1A1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF24 |