Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106856 |
| Name | oriT_CFBP 1156|unnamed |
| Organism | Xanthomonas hyacinthi strain CFBP 1156 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP043477 (43810..43904 [-], 95 nt) |
| oriT length | 95 nt |
| IRs (inverted repeats) | 26..33, 36..43 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 95 nt
>oriT_CFBP 1156|unnamed
CGAATAAGGGGATAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACCTATCCTGCCCGCCTTTCGGCGGTATGAACACCAAGGAAAG
CGAATAAGGGGATAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACCTATCCTGCCCGCCTTTCGGCGGTATGAACACCAAGGAAAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file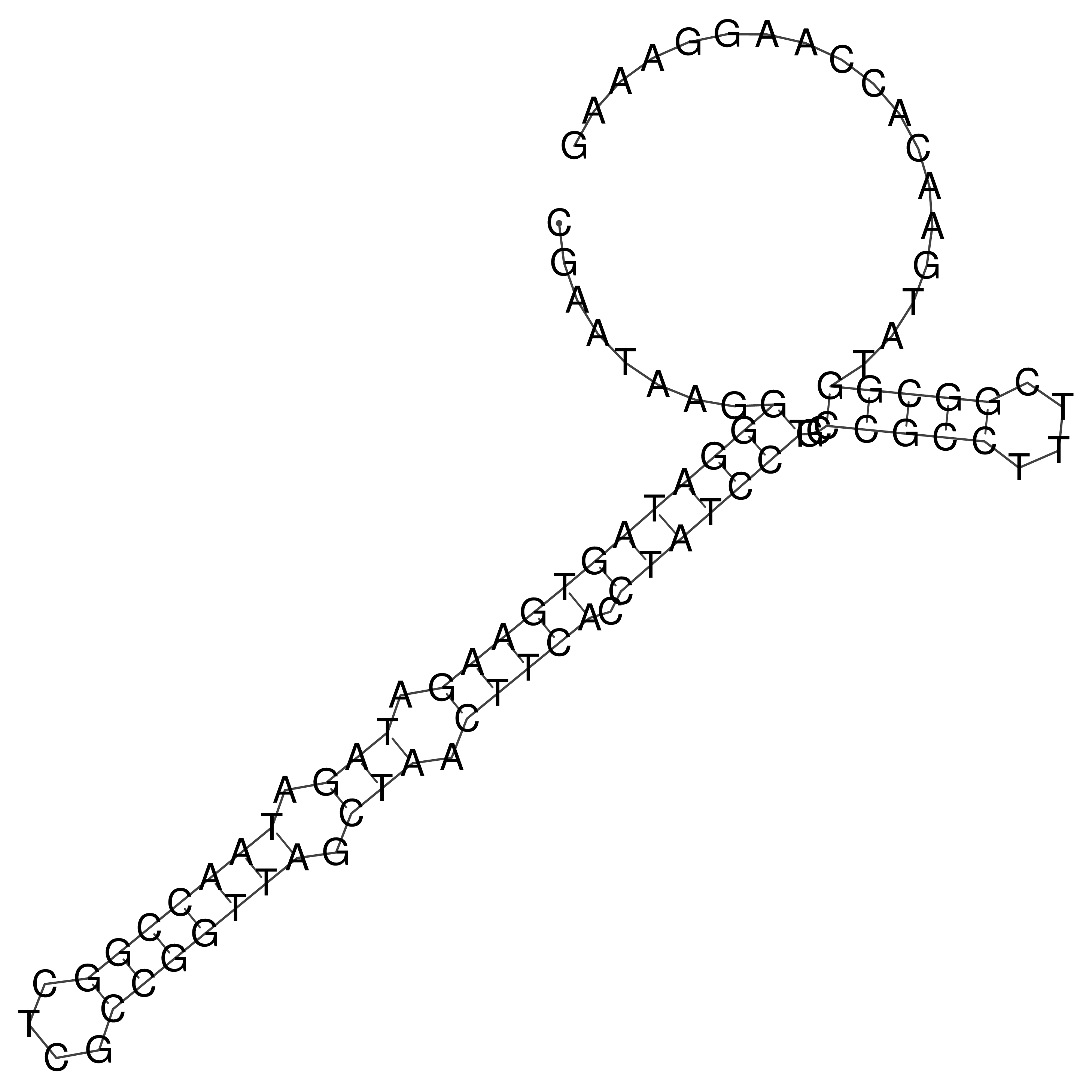
T4CP
| ID | 4588 | GenBank | WP_046977535 |
| Name | t4cp2_FZ025_RS21555_CFBP 1156|unnamed |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69719.95 Da Isoelectric Point: 9.0430
>WP_046977535.1 type IV secretory system conjugative DNA transfer family protein [Xanthomonas hyacinthi]
MTKDRFNNAVGPQVRAEKEGPGKVIPALAVLSLGGGLQAATQFFAHDFKYQAALGAHINHIYPPWGIVQW
AGKWYGQYPDAFMRAGSIGVTITGVGLISLVVAKMVLANSSKVNEYLHGSARWANVRDIQAAGLIPRARS
FWQTITGKDAAASSGVYVGAWIDKKGKQHYLRHNGPEHVLCYAPTRSGKGVGLVVPTLLSWGHSAVITDL
KGELWALTAGWRQKHAKNKVLRFEPATASGSVCWNPLDEIRLGTENEVGDVQNLATLIVDPDGKGLESHW
QKTSQALLVGVILHALYKTKAEGTPATLPGVDAMLADPNRDIGELWMEMTTYSHVDGQVHPVVGSAARDM
MDRPDEEAGSVLSTAKSYLALYRDPIVARNVSKSEFKIKDLMHHDNAVSLYIVTQPNDKARLRPLVRVMV
NMIVRLLADKMDFENGRPVAHYKHRMLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRE
TGYGHDESITSNCHVQNAYPPNRVETAEHLSKLTGQTTVAKEQITTSGRRTSALLGQVSRTIQEVQRPLL
TPDECLRMPGPVKTEGGDIAKAGDMVVYVAGYPAIYGKQPLYFQDPTFQARAAIPAPKNSDKLRNSFVAP
AGEGITI
MTKDRFNNAVGPQVRAEKEGPGKVIPALAVLSLGGGLQAATQFFAHDFKYQAALGAHINHIYPPWGIVQW
AGKWYGQYPDAFMRAGSIGVTITGVGLISLVVAKMVLANSSKVNEYLHGSARWANVRDIQAAGLIPRARS
FWQTITGKDAAASSGVYVGAWIDKKGKQHYLRHNGPEHVLCYAPTRSGKGVGLVVPTLLSWGHSAVITDL
KGELWALTAGWRQKHAKNKVLRFEPATASGSVCWNPLDEIRLGTENEVGDVQNLATLIVDPDGKGLESHW
QKTSQALLVGVILHALYKTKAEGTPATLPGVDAMLADPNRDIGELWMEMTTYSHVDGQVHPVVGSAARDM
MDRPDEEAGSVLSTAKSYLALYRDPIVARNVSKSEFKIKDLMHHDNAVSLYIVTQPNDKARLRPLVRVMV
NMIVRLLADKMDFENGRPVAHYKHRMLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRE
TGYGHDESITSNCHVQNAYPPNRVETAEHLSKLTGQTTVAKEQITTSGRRTSALLGQVSRTIQEVQRPLL
TPDECLRMPGPVKTEGGDIAKAGDMVVYVAGYPAIYGKQPLYFQDPTFQARAAIPAPKNSDKLRNSFVAP
AGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4589 | GenBank | WP_046977525 |
| Name | t4cp2_FZ025_RS21685_CFBP 1156|unnamed |
UniProt ID | _ |
| Length | 845 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 845 a.a. Molecular weight: 93736.73 Da Isoelectric Point: 6.3400
>WP_046977525.1 VirB4 family type IV secretion/conjugal transfer ATPase [Xanthomonas hyacinthi]
MIEAIAIGIAALGALLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAMVDDGVIVGKNGSFIASWL
YKGDDNASSTDEQREMVSFRINQALAGLGNGWMLHVDAVRRPAPNYTETGLSHFPDPVSAAMDEERRRLF
EGLGTMYEGYFIVTMTYFPPLLAQSKFVELMFDDEAKTPGNKARTQGLIDYFKRECNSIDSRLSSAVKLQ
RLRGNKVVNEDGSTVTHDDLLRWLQFCITGMDHPVLLPSNPMYLDAVIGGQELFGGVVPKIGRKFVQVVA
IEGFPLESTPGVLTALGELPVEYRWSSRFIFMDSHEAVKHLDKFRKKWKQKIRGFFDQVFNTNTGSVDQD
AMSMVNDAESAIAEVNSGLVAVGYYTSVVVLMDEDREKLETSARQVEKAINRLGFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGLNKAPCPMYPPLSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLALIAAQLRRYQGMSIYAFDKGMSMYPLAKATGGKHFTVAADDDKLAFCPLQFLES
KSDRAWAMEWIDTILALNNVETTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIRQYTVDGNM
GHLLDAEEDGLSLTDFTVFEIEELMNLGEKYALPTLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMGLNARQIE
ILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLETKFGDAWVHEWLAGRNLKLSD
YGVAA
MIEAIAIGIAALGALLLLILFARIRAVDAELKLKKHRSKDAGLADLLNYAAMVDDGVIVGKNGSFIASWL
YKGDDNASSTDEQREMVSFRINQALAGLGNGWMLHVDAVRRPAPNYTETGLSHFPDPVSAAMDEERRRLF
EGLGTMYEGYFIVTMTYFPPLLAQSKFVELMFDDEAKTPGNKARTQGLIDYFKRECNSIDSRLSSAVKLQ
RLRGNKVVNEDGSTVTHDDLLRWLQFCITGMDHPVLLPSNPMYLDAVIGGQELFGGVVPKIGRKFVQVVA
IEGFPLESTPGVLTALGELPVEYRWSSRFIFMDSHEAVKHLDKFRKKWKQKIRGFFDQVFNTNTGSVDQD
AMSMVNDAESAIAEVNSGLVAVGYYTSVVVLMDEDREKLETSARQVEKAINRLGFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGLNKAPCPMYPPLSPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLALIAAQLRRYQGMSIYAFDKGMSMYPLAKATGGKHFTVAADDDKLAFCPLQFLES
KSDRAWAMEWIDTILALNNVETTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIRQYTVDGNM
GHLLDAEEDGLSLTDFTVFEIEELMNLGEKYALPTLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMGLNARQIE
ILATAIPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLETKFGDAWVHEWLAGRNLKLSD
YGVAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 19179..29788
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FZ025_RS21600 (FZ025_21760) | 14789..15157 | + | 369 | WP_146093616 | hypothetical protein | - |
| FZ025_RS22625 | 15261..15422 | - | 162 | Protein_12 | antitoxin VbhA family protein | - |
| FZ025_RS21610 (FZ025_21765) | 15389..15670 | + | 282 | Protein_13 | helix-turn-helix domain-containing protein | - |
| FZ025_RS21615 (FZ025_21770) | 15627..16034 | - | 408 | WP_031944013 | hypothetical protein | - |
| FZ025_RS21620 (FZ025_21775) | 16031..16516 | - | 486 | WP_015063510 | thermonuclease family protein | - |
| FZ025_RS21625 (FZ025_21780) | 16518..16943 | - | 426 | WP_053057113 | OmpA family protein | - |
| FZ025_RS21630 (FZ025_21785) | 16957..17655 | - | 699 | WP_244292574 | TraX family protein | - |
| FZ025_RS21635 (FZ025_21790) | 17658..17921 | - | 264 | WP_046977531 | hypothetical protein | - |
| FZ025_RS21640 (FZ025_21795) | 17947..18552 | - | 606 | WP_046977530 | transglycosylase SLT domain-containing protein | - |
| FZ025_RS21645 (FZ025_21800) | 18566..19159 | - | 594 | WP_015063527 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| FZ025_RS21650 (FZ025_21805) | 19179..20924 | - | 1746 | WP_053057112 | P-type conjugative transfer protein TrbL | virB6 |
| FZ025_RS21655 (FZ025_21810) | 20935..21174 | - | 240 | WP_031944010 | entry exclusion lipoprotein TrbK | - |
| FZ025_RS21660 (FZ025_21815) | 21185..21958 | - | 774 | WP_104558932 | P-type conjugative transfer protein TrbJ | virB5 |
| FZ025_RS21665 (FZ025_21820) | 21977..23371 | - | 1395 | WP_046977528 | TrbI/VirB10 family protein | virB10 |
| FZ025_RS21670 (FZ025_21825) | 23377..23844 | - | 468 | WP_046977527 | conjugal transfer protein TrbH | - |
| FZ025_RS21675 (FZ025_21830) | 23847..24752 | - | 906 | WP_046977526 | P-type conjugative transfer protein TrbG | virB9 |
| FZ025_RS21680 (FZ025_21835) | 24765..25541 | - | 777 | WP_015063521 | conjugal transfer protein TrbF | virB8 |
| FZ025_RS21685 (FZ025_21840) | 25538..28075 | - | 2538 | WP_046977525 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| FZ025_RS21690 (FZ025_21845) | 28072..28383 | - | 312 | WP_011171720 | conjugal transfer protein TrbD | virB3 |
| FZ025_RS21695 (FZ025_21850) | 28386..28814 | - | 429 | WP_011171719 | IncP-type conjugal transfer pilin TrbC | virB2 |
| FZ025_RS21700 (FZ025_21855) | 28826..29788 | - | 963 | WP_011171718 | P-type conjugative transfer ATPase TrbB | virB11 |
| FZ025_RS21705 (FZ025_21860) | 30043..30420 | - | 378 | WP_031944007 | transcriptional regulator | - |
| FZ025_RS21710 (FZ025_21865) | 30741..31610 | + | 870 | WP_031944006 | plasmid replication initiator TrfA | - |
| FZ025_RS21715 (FZ025_21875) | 32856..33140 | + | 285 | WP_031944038 | hypothetical protein | - |
| FZ025_RS21720 (FZ025_21880) | 33238..33672 | + | 435 | WP_011171688 | antirestriction protein | - |
Host bacterium
| ID | 7293 | GenBank | NZ_CP043477 |
| Plasmid name | CFBP 1156|unnamed | Incompatibility group | - |
| Plasmid size | 44381 bp | Coordinate of oriT [Strand] | 43810..43904 [-] |
| Host baterium | Xanthomonas hyacinthi strain CFBP 1156 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9, AcrIF24 |