Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106855 |
| Name | oriT_SRCM103227|unnamed1 |
| Organism | Mixta theicola strain SRCM103227 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP028274 (51597..51683 [-], 87 nt) |
| oriT length | 87 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 87 nt
>oriT_SRCM103227|unnamed1
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTAATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file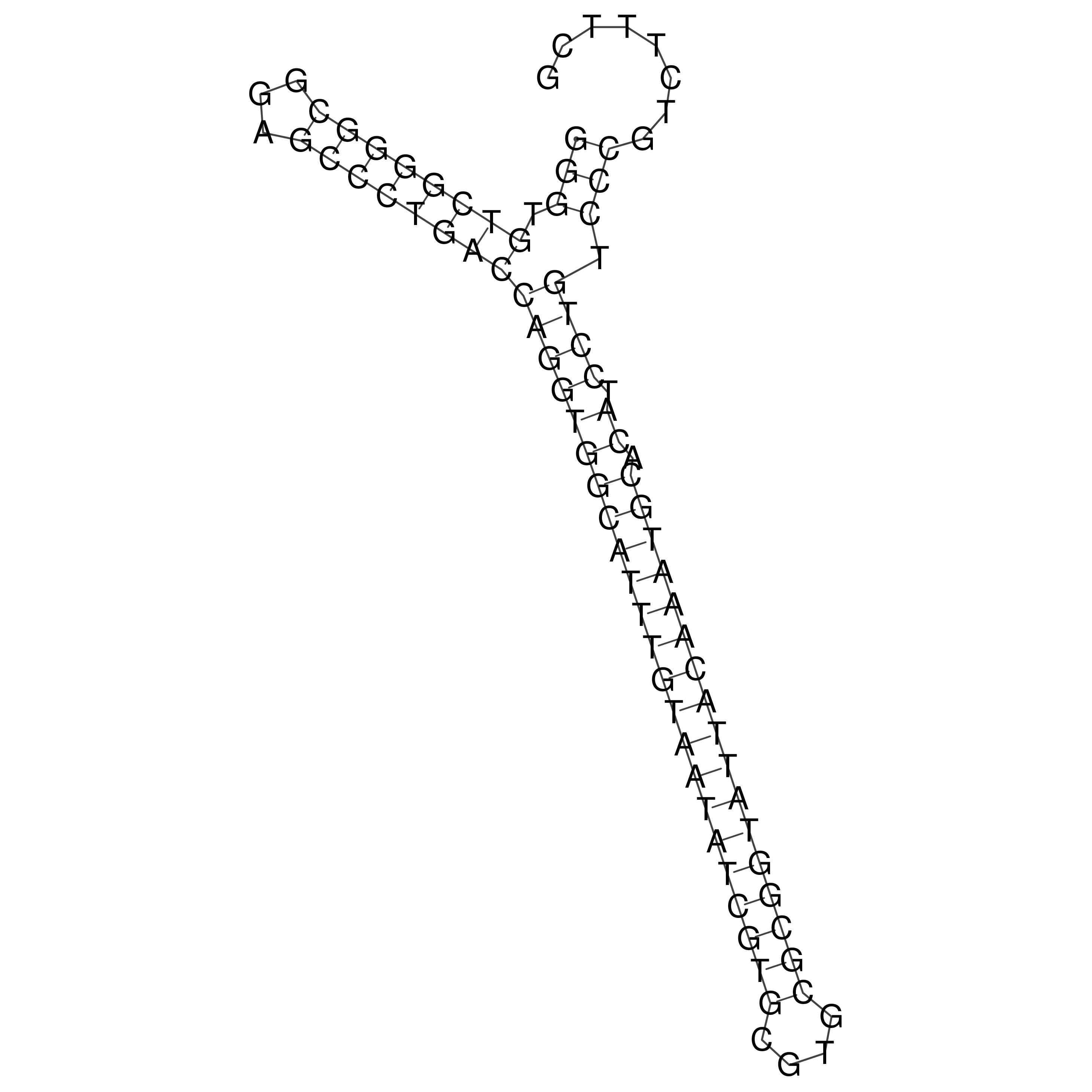
Relaxase
| ID | 4523 | GenBank | WP_160251420 |
| Name | Relaxase_C7M52_RS21205_SRCM103227|unnamed1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102392.61 Da Isoelectric Point: 8.2772
>WP_160251420.1 relaxase/mobilization nuclease domain-containing protein [Mixta theicola]
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVKASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNHIVRRTSVERDRQSAWKRGRNQSLREYIADTAIAGLRESPVTDWPSLHQRFAKEGLFLTKE
SGELKVKDGWERERPGVTLSAFGHAFDTDKLIRKLGEFVPPAQDIFTQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYAIARLRAPLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDSTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKNIFQQVPPAEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIEMLIGSGEFTWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
SEPHAGSFVSVPADIFERVKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQAIHDECRMRKARIRIEHRDPLVRKLHYHIAELQRMQALIALKETMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDNRCVVICEPGGTPMYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNSGDYDALKRDMVKVAPVLFSRSPSVAMAPEGDNEQFNTAFAKM
VAWHNVNHRDEGEYQISRPEVDQVRVESEMRFAEMTNQGSNTEAVRNESGPNWKPPSPL
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVKASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKAKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNHIVRRTSVERDRQSAWKRGRNQSLREYIADTAIAGLRESPVTDWPSLHQRFAKEGLFLTKE
SGELKVKDGWERERPGVTLSAFGHAFDTDKLIRKLGEFVPPAQDIFTQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYAIARLRAPLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDSTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKNIFQQVPPAEQFTGGGLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIEMLIGSGEFTWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
SEPHAGSFVSVPADIFERVKPVSRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQAIHDECRMRKARIRIEHRDPLVRKLHYHIAELQRMQALIALKETMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDNRCVVICEPGGTPMYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNSGDYDALKRDMVKVAPVLFSRSPSVAMAPEGDNEQFNTAFAKM
VAWHNVNHRDEGEYQISRPEVDQVRVESEMRFAEMTNQGSNTEAVRNESGPNWKPPSPL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 2023 | GenBank | WP_160251419 |
| Name | WP_160251419_SRCM103227|unnamed1 |
UniProt ID | _ |
| Length | 111 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 111 a.a. Molecular weight: 12663.63 Da Isoelectric Point: 10.3118
>WP_160251419.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacter]
MSDKASRRGSDKRQKTVIKTLRFTPDEIEEIEQKAEAVGQSVSAYIRNAAMNRKMISLIDDSFMTELLRL
GRLQKHLFVEGKRVGDKEYSTVLVAITELSTTLRKKLMSND
MSDKASRRGSDKRQKTVIKTLRFTPDEIEEIEQKAEAVGQSVSAYIRNAAMNRKMISLIDDSFMTELLRL
GRLQKHLFVEGKRVGDKEYSTVLVAITELSTTLRKKLMSND
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 4587 | GenBank | WP_160251421 |
| Name | trbC_C7M52_RS21220_SRCM103227|unnamed1 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87607.89 Da Isoelectric Point: 6.8632
>WP_160251421.1 F-type conjugative transfer protein TrbC [Mixta theicola]
MNYTPVNQALMKRNAWNSPVLELLRDHLLYAGIMAGSVVAGFICPLSIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKIDPSEDRKVRRSLFRRFPSLFQYETTQESKGKGIFYLGYRRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IHNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAELYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHRLE
GLDSEALEITKYKGPFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIVMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTITNAAGQEARANMVSLQRRDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKAAKAEPWKIIDTFQQRLANRQEAHNLMTEYDTDHRGREENLWEEALEVIRTTTMAERTI
RYITLNKPESEPTQVSAEEIQLAPDAILRTLTLPQPAYIPPSRYRNEPFISPKLPDSPHPDMEWPE
MNYTPVNQALMKRNAWNSPVLELLRDHLLYAGIMAGSVVAGFICPLSIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKIDPSEDRKVRRSLFRRFPSLFQYETTQESKGKGIFYLGYRRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IHNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAELYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHRLE
GLDSEALEITKYKGPFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIVMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTITNAAGQEARANMVSLQRRDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKAAKAEPWKIIDTFQQRLANRQEAHNLMTEYDTDHRGREENLWEEALEVIRTTTMAERTI
RYITLNKPESEPTQVSAEEIQLAPDAILRTLTLPQPAYIPPSRYRNEPFISPKLPDSPHPDMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 57806..80821
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| C7M52_RS21210 (C7M52_04190) | 54822..55025 | - | 204 | WP_133147680 | hypothetical protein | - |
| C7M52_RS21215 (C7M52_04191) | 55159..55428 | - | 270 | WP_022650007 | hypothetical protein | - |
| C7M52_RS21220 (C7M52_04192) | 55502..57802 | - | 2301 | WP_160251421 | F-type conjugative transfer protein TrbC | - |
| C7M52_RS21225 (C7M52_04193) | 57806..59062 | - | 1257 | WP_160251422 | thioredoxin fold domain-containing protein | trbB |
| C7M52_RS21230 (C7M52_04194) | 59043..60251 | - | 1209 | WP_032650374 | hypothetical protein | trbA |
| C7M52_RS21235 (C7M52_04195) | 60572..61204 | - | 633 | WP_155026669 | plasmid IncI1-type surface exclusion protein ExcA | - |
| C7M52_RS21240 (C7M52_04196) | 61266..63425 | - | 2160 | WP_047625721 | DotA/TraY family protein | traY |
| C7M52_RS21245 (C7M52_04197) | 63483..64025 | - | 543 | WP_160251423 | conjugal transfer protein TraX | - |
| C7M52_RS21250 (C7M52_04198) | 64022..65224 | - | 1203 | WP_032650382 | conjugal transfer protein TraW | traW |
| C7M52_RS21255 (C7M52_04200) | 65806..68859 | - | 3054 | WP_022649998 | hypothetical protein | traU |
| C7M52_RS21260 (C7M52_04201) | 68946..69671 | - | 726 | WP_244316988 | conjugal transfer protein TraT | traT |
| C7M52_RS21265 (C7M52_04202) | 69855..70259 | - | 405 | WP_095427107 | DUF6750 family protein | traR |
| C7M52_RS21270 (C7M52_04203) | 70293..70817 | - | 525 | WP_047625719 | conjugal transfer protein TraQ | traQ |
| C7M52_RS21275 (C7M52_04204) | 70817..71590 | - | 774 | WP_123839398 | conjugal transfer protein TraP | traP |
| C7M52_RS21280 (C7M52_04205) | 71571..72878 | - | 1308 | WP_095427106 | conjugal transfer protein TraO | traO |
| C7M52_RS21285 (C7M52_04206) | 72878..73858 | - | 981 | WP_160251424 | DotH/IcmK family type IV secretion protein | traN |
| C7M52_RS21290 (C7M52_04207) | 73869..74570 | - | 702 | WP_095427104 | DotI/IcmL family type IV secretion protein | traM |
| C7M52_RS21295 (C7M52_04208) | 74576..74926 | - | 351 | WP_022649990 | hypothetical protein | traL |
| C7M52_RS21300 (C7M52_04209) | 74944..78486 | - | 3543 | WP_160251425 | LPD7 domain-containing protein | - |
| C7M52_RS21305 (C7M52_04210) | 78558..78845 | - | 288 | WP_022649988 | hypothetical protein | traK |
| C7M52_RS21310 (C7M52_04211) | 78842..80002 | - | 1161 | WP_225828800 | plasmid transfer ATPase TraJ | virB11 |
| C7M52_RS21315 (C7M52_04212) | 80015..80821 | - | 807 | WP_160251426 | type IV secretory system conjugative DNA transfer family protein | traI |
| C7M52_RS21320 (C7M52_04213) | 80818..81291 | - | 474 | WP_032650400 | DotD/TraH family lipoprotein | - |
| C7M52_RS21325 (C7M52_04214) | 81361..81777 | - | 417 | WP_160251427 | hypothetical protein | - |
Host bacterium
| ID | 7292 | GenBank | NZ_CP028274 |
| Plasmid name | SRCM103227|unnamed1 | Incompatibility group | - |
| Plasmid size | 82504 bp | Coordinate of oriT [Strand] | 51597..51683 [-] |
| Host baterium | Mixta theicola strain SRCM103227 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |