Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106784 |
| Name | oriT_pI9455333_MCR10 |
| Organism | Enterobacter ludwigii strain I9455333cz |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP122443 (94594..94642 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pI9455333_MCR10
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file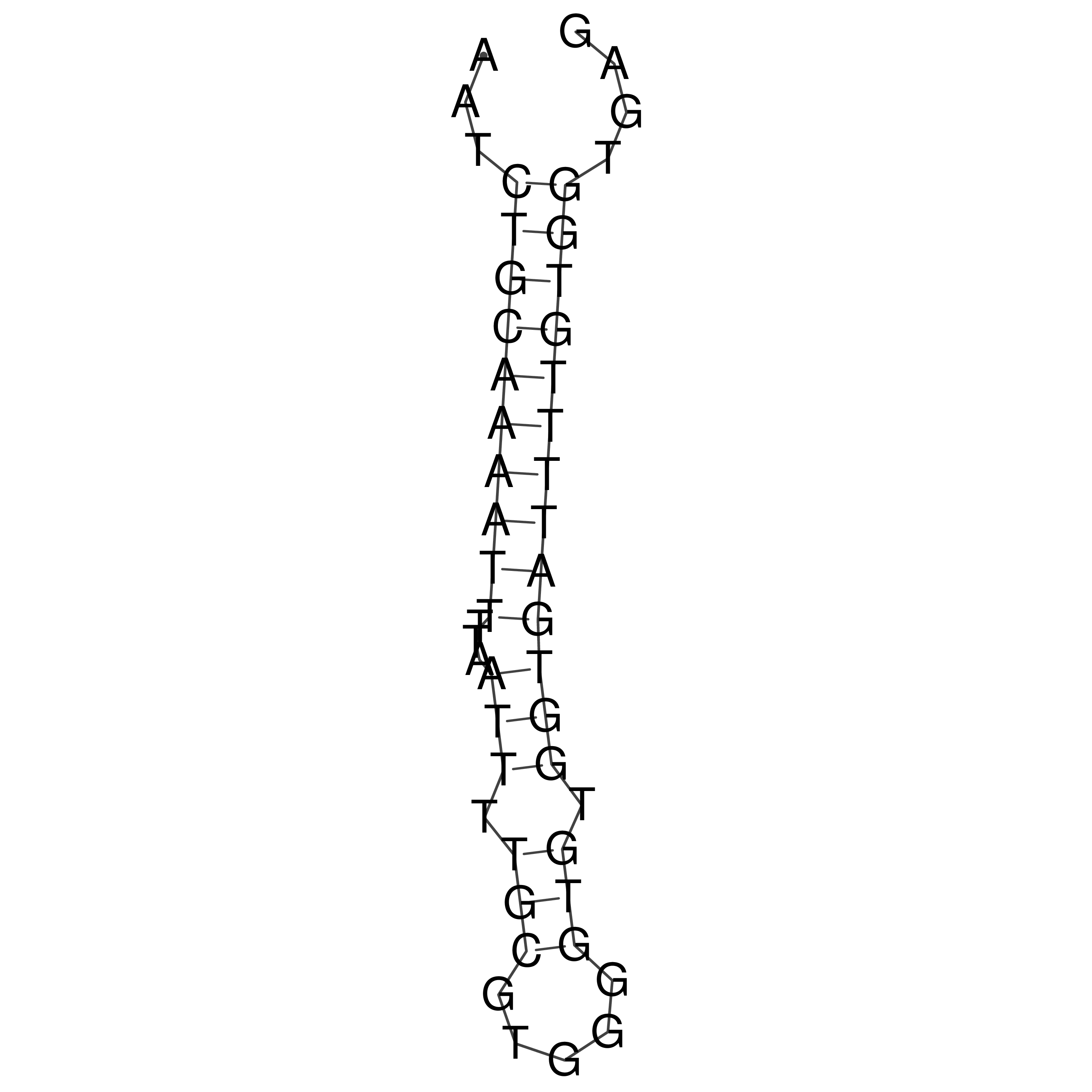
Relaxase
| ID | 4473 | GenBank | WP_023332871 |
| Name | traI_QCL67_RS26335_pI9455333_MCR10 |
UniProt ID | _ |
| Length | 1751 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1751 a.a. Molecular weight: 191031.21 Da Isoelectric Point: 6.0768
>WP_023332871.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGKAGDYYTKEDNYYVIGSMDERWQGKGAEALGLEGKTVDKALFTELLKGKLPDGSDLSRI
QDGMNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIEAHNQAVTEAIRQVETLAATRVMVDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREVLKPLVE
GLGYETEVVGKHGMWEMKDVPVEPFSTRTQEINAAAGPDASLKSRDVATLDTRKAKETLDPAQKVTEWLS
TLKETGFNLKEYREAADQRAKTQREVPSPEAPAGPNVDDIVTQAISGLSDRKVQFTYTELLARTVSQMEA
RPGVIETARAGIEAAIGREQLIPLDREKGIFTSGIHVLDELSVRALSQEVLRQNHVSMVPEKATARQTSF
SDAVTVLAQDRPAIGIVSGQGGAAGQRERVTEMAMMAREQGRDVHIIAADNRSLEFLLKEEKLAGEIITG
KSALQDGTAFVPGGTLIIDQAEKLSLKETINLLDGAMRNNVQILMSDSGKRSGTGSALTVMKESGVTAYK
WQGGKQTTASVISEPDRNQRFTRMASDFVASVRDGQESVVQIRGPREQGILTGLIRENLRNEGLIGSHEE
TITALTPVWLDSKNRGIRDNYREGMVMERWDPEKRIHDRFIIDRVTAVSNTLTLKDEKGESLNLKVSAVD
NTWTLFRPQQLPVAEGERLAILGKIPETRLKGGDTVQVLKAENGVLTVQHPGQKTTQKLAAGNTPFTGIK
VGHGWVENPGRSVSETATVFASLSQRELDNATLNALAQSGSQVRLYSAQNEQKTVEKLGRHTAFSVVTEQ
LKTRAGEQDISAAIAHQKSQLHTPAEQAIHLAVPSLESDKLTFTRPQLLVAAHEFSDGTVPFSDIEKTIG
EQIKSGDLLHVPVAAGHGNDLLISRQTWDAEKSLLTMVLEGKNAVTPLMAKVPGSLMEGLTSGQRNATRM
ILETRDRFTLVQGYAGVGKTTQFRAVMSAVNLLPEDSRPKVIGLGPTHRAVGEMQSAGVEAQTTASFLHD
TQLLMRSGQTPDFSNTLFLVDESSMLGLSDTAKAMSLIAAGGGRAVSSGDTDQLQSIAPGQPFRLMQKRS
AADVAIMQEIVRQVPELRPAVYSLIDRDIHSALSTIERVEPAQVPRKENAWVPESSVMEFTPEQEKAIQK
AIGEGEVLPDISPVSLYDAIVRDYTGRTESAQAQTLIITHLNDDRRVLNRMIHDEKMRNGELGEDSVTLP
VLVTSNIRDGELRKVSTWEAHKGAIALVDNQYHQIVGVSKADQLITLRSDEGTERLISPREASAEGVTLY
RQESITVSPGDRMRFSKSDTERGYVANSVWRVESISGDSVTLNDGKSTRTVTPAADKAEAHVDLAYAITT
HGAQGASEPFSIELEGVTGARERMVSFESAYVALSRMKQHTQVYTDDRKGWTDAVSRSQEKASAHDVLEP
LNDRAVKSAEQLFGRAIPLTESPAGRAALQQSGIARGQSTGKFISPGKKYPLPHVALPAFDRNGKSAGIW
LSPLTDKDGRLQAIGGEGRVMGNEEARFVALQGSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPGAITGGRVWGDIDAVVTGGSGANIPLPPDLLAQKTAEELQRKQIEKQAQETARELTGEKRKQDEVGE
QIKDIITDVVRGLERQKGQHERFQLPDEPENRLRNEAVQKVVADNQQQDRLQQIERDMVRDLTREKTLGG
D
MMSIGSVKSAGKAGDYYTKEDNYYVIGSMDERWQGKGAEALGLEGKTVDKALFTELLKGKLPDGSDLSRI
QDGMNKHRPGYDLTFSAPKSVSVMAMLGGDKRLIEAHNQAVTEAIRQVETLAATRVMVDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYANQIAFGKIYREVLKPLVE
GLGYETEVVGKHGMWEMKDVPVEPFSTRTQEINAAAGPDASLKSRDVATLDTRKAKETLDPAQKVTEWLS
TLKETGFNLKEYREAADQRAKTQREVPSPEAPAGPNVDDIVTQAISGLSDRKVQFTYTELLARTVSQMEA
RPGVIETARAGIEAAIGREQLIPLDREKGIFTSGIHVLDELSVRALSQEVLRQNHVSMVPEKATARQTSF
SDAVTVLAQDRPAIGIVSGQGGAAGQRERVTEMAMMAREQGRDVHIIAADNRSLEFLLKEEKLAGEIITG
KSALQDGTAFVPGGTLIIDQAEKLSLKETINLLDGAMRNNVQILMSDSGKRSGTGSALTVMKESGVTAYK
WQGGKQTTASVISEPDRNQRFTRMASDFVASVRDGQESVVQIRGPREQGILTGLIRENLRNEGLIGSHEE
TITALTPVWLDSKNRGIRDNYREGMVMERWDPEKRIHDRFIIDRVTAVSNTLTLKDEKGESLNLKVSAVD
NTWTLFRPQQLPVAEGERLAILGKIPETRLKGGDTVQVLKAENGVLTVQHPGQKTTQKLAAGNTPFTGIK
VGHGWVENPGRSVSETATVFASLSQRELDNATLNALAQSGSQVRLYSAQNEQKTVEKLGRHTAFSVVTEQ
LKTRAGEQDISAAIAHQKSQLHTPAEQAIHLAVPSLESDKLTFTRPQLLVAAHEFSDGTVPFSDIEKTIG
EQIKSGDLLHVPVAAGHGNDLLISRQTWDAEKSLLTMVLEGKNAVTPLMAKVPGSLMEGLTSGQRNATRM
ILETRDRFTLVQGYAGVGKTTQFRAVMSAVNLLPEDSRPKVIGLGPTHRAVGEMQSAGVEAQTTASFLHD
TQLLMRSGQTPDFSNTLFLVDESSMLGLSDTAKAMSLIAAGGGRAVSSGDTDQLQSIAPGQPFRLMQKRS
AADVAIMQEIVRQVPELRPAVYSLIDRDIHSALSTIERVEPAQVPRKENAWVPESSVMEFTPEQEKAIQK
AIGEGEVLPDISPVSLYDAIVRDYTGRTESAQAQTLIITHLNDDRRVLNRMIHDEKMRNGELGEDSVTLP
VLVTSNIRDGELRKVSTWEAHKGAIALVDNQYHQIVGVSKADQLITLRSDEGTERLISPREASAEGVTLY
RQESITVSPGDRMRFSKSDTERGYVANSVWRVESISGDSVTLNDGKSTRTVTPAADKAEAHVDLAYAITT
HGAQGASEPFSIELEGVTGARERMVSFESAYVALSRMKQHTQVYTDDRKGWTDAVSRSQEKASAHDVLEP
LNDRAVKSAEQLFGRAIPLTESPAGRAALQQSGIARGQSTGKFISPGKKYPLPHVALPAFDRNGKSAGIW
LSPLTDKDGRLQAIGGEGRVMGNEEARFVALQGSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPGAITGGRVWGDIDAVVTGGSGANIPLPPDLLAQKTAEELQRKQIEKQAQETARELTGEKRKQDEVGE
QIKDIITDVVRGLERQKGQHERFQLPDEPENRLRNEAVQKVVADNQQQDRLQQIERDMVRDLTREKTLGG
D
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1990 | GenBank | WP_001568107 |
| Name | WP_001568107_pI9455333_MCR10 |
UniProt ID | _ |
| Length | 128 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 128 a.a. Molecular weight: 14784.78 Da Isoelectric Point: 4.6362
>WP_001568107.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacteriaceae]
MARVQAYPSDEVVDKINAIVEKRRAEGAKEKDISFSSVATMLLELGLRVYEAQMERKESGFNQKEFNKVL
LENVMKTQFTISKVLAMDSLSPHINGDDRFDFKSMVMNIRDDAREVVERFFPSQDEEE
MARVQAYPSDEVVDKINAIVEKRRAEGAKEKDISFSSVATMLLELGLRVYEAQMERKESGFNQKEFNKVL
LENVMKTQFTISKVLAMDSLSPHINGDDRFDFKSMVMNIRDDAREVVERFFPSQDEEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4521 | GenBank | WP_023332870 |
| Name | traD_QCL67_RS26330_pI9455333_MCR10 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 82700.58 Da Isoelectric Point: 5.9222
>WP_023332870.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIASMRFRMFGQIANIILYVLFLFFWVLCGLILMYRLSWQTFVNGAVYWWCTTLGPMR
DIIRSQPVYTINYYGQQLQYTSEQILKDKYTIWCGEQLWTGFVFAGTVSLIICIVAFFVASWVLGHQGKQ
QSEDEVTGGRQLSEKPKEVARKMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSIDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRSVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKGIA
EFAAGEIGEKEIKKASENYSYGADPVRDGVSTGKEQKRETIVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRQLNQAIDDKLAAVLAAREAEGQSARILFMPDPVETVPVEKTDEKPASPVAAVPTP
QEDKAKVPPVTASNTFHKPASAAAAAASASVTQAGGVEQELHEKPEEQLPLPPGVNKDGEIEDMNAWDEW
QSSSDVLRDMHRREEVNINHSHHVDRDDINLGSNF
MSFNAKDMTQGGQIASMRFRMFGQIANIILYVLFLFFWVLCGLILMYRLSWQTFVNGAVYWWCTTLGPMR
DIIRSQPVYTINYYGQQLQYTSEQILKDKYTIWCGEQLWTGFVFAGTVSLIICIVAFFVASWVLGHQGKQ
QSEDEVTGGRQLSEKPKEVARKMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSIDKILNPLDSRCAAWDLWKECLTLPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRSVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKGIA
EFAAGEIGEKEIKKASENYSYGADPVRDGVSTGKEQKRETIVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRQLNQAIDDKLAAVLAAREAEGQSARILFMPDPVETVPVEKTDEKPASPVAAVPTP
QEDKAKVPPVTASNTFHKPASAAAAAASASVTQAGGVEQELHEKPEEQLPLPPGVNKDGEIEDMNAWDEW
QSSSDVLRDMHRREEVNINHSHHVDRDDINLGSNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 94035..120881
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| QCL67_RS26140 (QCL67_26140) | 89088..89375 | + | 288 | WP_000323025 | type II toxin-antitoxin system mRNA interferase RelE | - |
| QCL67_RS26145 (QCL67_26145) | 89426..90460 | - | 1035 | Protein_107 | IS481 family transposase | - |
| QCL67_RS26585 | 90561..90623 | + | 63 | Protein_108 | hypothetical protein | - |
| QCL67_RS26590 | 90595..90693 | + | 99 | Protein_109 | Hok/Gef family protein | - |
| QCL67_RS26155 (QCL67_26155) | 90788..91996 | + | 1209 | WP_015386429 | IS256 family transposase | - |
| QCL67_RS26160 (QCL67_26160) | 92278..92409 | - | 132 | WP_255344245 | hypothetical protein | - |
| QCL67_RS26170 (QCL67_26170) | 92807..93628 | + | 822 | WP_023332864 | DUF932 domain-containing protein | - |
| QCL67_RS26175 (QCL67_26175) | 93694..94002 | + | 309 | WP_023332865 | DUF5983 family protein | - |
| QCL67_RS26180 (QCL67_26180) | 94035..94520 | - | 486 | WP_032629900 | transglycosylase SLT domain-containing protein | virB1 |
| QCL67_RS26185 (QCL67_26185) | 94943..95329 | + | 387 | WP_001568107 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| QCL67_RS26190 (QCL67_26190) | 95549..96238 | + | 690 | WP_001568106 | helix-turn-helix domain-containing protein | - |
| QCL67_RS26195 (QCL67_26195) | 96413..96577 | + | 165 | WP_071594636 | TraY domain-containing protein | - |
| QCL67_RS26200 (QCL67_26200) | 96656..97024 | + | 369 | WP_001568105 | type IV conjugative transfer system pilin TraA | - |
| QCL67_RS26205 (QCL67_26205) | 97104..97472 | + | 369 | WP_001568104 | type IV conjugative transfer system pilin TraA | - |
| QCL67_RS26210 (QCL67_26210) | 97486..97791 | + | 306 | WP_024191986 | type IV conjugative transfer system protein TraL | traL |
| QCL67_RS26215 (QCL67_26215) | 97810..98376 | + | 567 | WP_001568102 | type IV conjugative transfer system protein TraE | traE |
| QCL67_RS26220 (QCL67_26220) | 98363..99103 | + | 741 | WP_001568101 | type-F conjugative transfer system secretin TraK | traK |
| QCL67_RS26225 (QCL67_26225) | 99103..100515 | + | 1413 | WP_001568100 | F-type conjugal transfer pilus assembly protein TraB | traB |
| QCL67_RS26230 (QCL67_26230) | 100508..100744 | + | 237 | WP_001568098 | hypothetical protein | virb4 |
| QCL67_RS26235 (QCL67_26235) | 100763..101332 | + | 570 | WP_001568094 | type IV conjugative transfer system lipoprotein TraV | traV |
| QCL67_RS26240 (QCL67_26240) | 101446..101859 | + | 414 | WP_001568093 | hypothetical protein | - |
| QCL67_RS26245 (QCL67_26245) | 101864..102262 | + | 399 | WP_001568092 | hypothetical protein | - |
| QCL67_RS26250 (QCL67_26250) | 102725..105364 | + | 2640 | WP_001568090 | type IV secretion system protein TraC | virb4 |
| QCL67_RS26255 (QCL67_26255) | 105364..105741 | + | 378 | WP_001568089 | type-F conjugative transfer system protein TrbI | - |
| QCL67_RS26260 (QCL67_26260) | 105738..106367 | + | 630 | WP_023332867 | type-F conjugative transfer system protein TraW | traW |
| QCL67_RS26265 (QCL67_26265) | 106399..107370 | + | 972 | WP_176602138 | conjugal transfer pilus assembly protein TraU | traU |
| QCL67_RS26270 (QCL67_26270) | 107383..108009 | + | 627 | WP_001568086 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| QCL67_RS26275 (QCL67_26275) | 108006..109898 | + | 1893 | WP_001568085 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| QCL67_RS26280 (QCL67_26280) | 109911..110171 | + | 261 | WP_001568084 | conjugal transfer protein TrbE | - |
| QCL67_RS26285 (QCL67_26285) | 110168..110500 | + | 333 | WP_001568083 | hypothetical protein | - |
| QCL67_RS26290 (QCL67_26290) | 110516..111265 | + | 750 | WP_001568082 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| QCL67_RS26295 (QCL67_26295) | 111276..111509 | + | 234 | WP_001568081 | type-F conjugative transfer system pilin chaperone TraQ | - |
| QCL67_RS26300 (QCL67_26300) | 111529..112065 | + | 537 | WP_032160235 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| QCL67_RS26305 (QCL67_26305) | 112052..113422 | + | 1371 | WP_023332869 | conjugal transfer pilus assembly protein TraH | traH |
| QCL67_RS26310 (QCL67_26310) | 113422..116271 | + | 2850 | WP_001568078 | conjugal transfer mating-pair stabilization protein TraG | traG |
| QCL67_RS26315 (QCL67_26315) | 116274..116468 | + | 195 | WP_250128443 | hypothetical protein | - |
| QCL67_RS26325 (QCL67_26325) | 117758..118489 | + | 732 | WP_001568076 | conjugal transfer complement resistance protein TraT | - |
| QCL67_RS26330 (QCL67_26330) | 118674..120881 | + | 2208 | WP_023332870 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 7221 | GenBank | NZ_CP122443 |
| Plasmid name | pI9455333_MCR10 | Incompatibility group | IncFIA |
| Plasmid size | 129863 bp | Coordinate of oriT [Strand] | 94594..94642 [-] |
| Host baterium | Enterobacter ludwigii strain I9455333cz |
Cargo genes
| Drug resistance gene | mcr-10 |
| Virulence gene | mrkA, mrkB, mrkC, mrkF |
| Metal resistance gene | ncrY, ncrC, ncrB, ncrA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |