Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106750 |
| Name | oriT_ZM13|pD |
| Organism | Ancylobacter polymorphus strain ZM13 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP083243 (26553..26607 [-], 55 nt) |
| oriT length | 55 nt |
| IRs (inverted repeats) | 22..28, 33..39 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 55 nt
>oriT_ZM13|pD
CCCCGCCAGGGCCGGCGCAGGACGTCGCGGCAGCGACGTATTACTGCGCCCTTGG
CCCCGCCAGGGCCGGCGCAGGACGTCGCGGCAGCGACGTATTACTGCGCCCTTGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file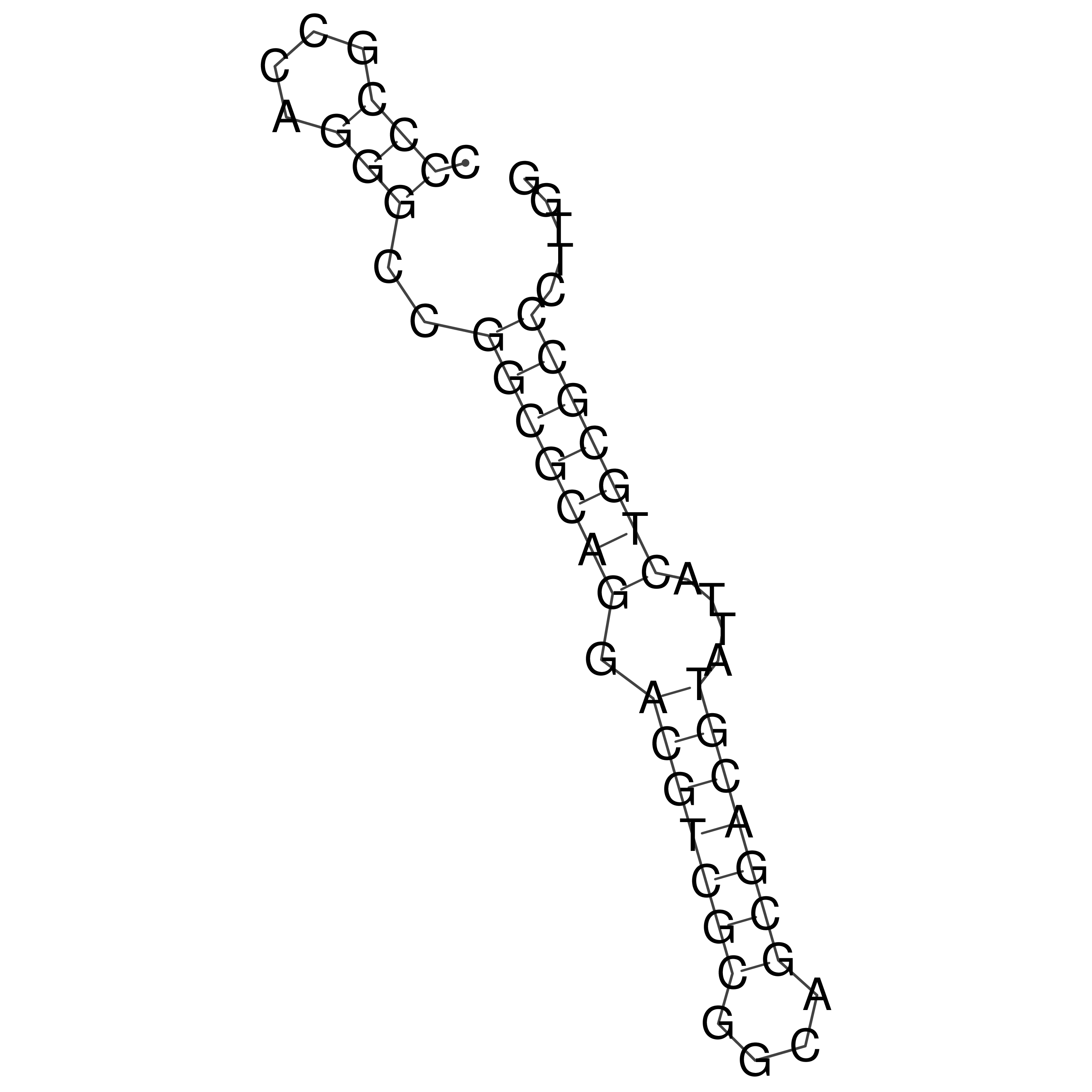
Relaxase
| ID | 4451 | GenBank | WP_244451514 |
| Name | traA_K9D25_RS24490_ZM13|pD |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123151.40 Da Isoelectric Point: 10.0859
>WP_244451514.1 Ti-type conjugative transfer relaxase TraA [Ancylobacter polymorphus]
MAITHFTPQLIGRGNGRSAVLSAAYRHCARMEYEAEARTVDYSNKRNLAHEEFLLPADAPEWARALIADR
SVAGAVEAFWNRVEAFEKREDAQFGREFIIALPVELSKEQNIALMRQFVLEQVLARGQVADWVYHDEPGN
PHVHLMTTLRPLTEDGFGPKRIPVIGEDGEVLRNKAGKIVYRLWSGEKTEFLEQRNGWLDLQNQHLALAG
LEIRVDGRSYAERGIDLVPTTHIGVGAKAIQRKSTREGEAVDLERLKLFEAQRTENRRRILLRPEIVLDL
LSSEKSVFDQRDIVKVLHRYVDDAGTFQQLMARILQSPELLRIERESVDFATGERLPARYTTRELIRVEA
GMARQAIWLSGRSSHGVRERVLEQVFARHERLSEEQRTAIEHVTKPGGIAAVVGRAGAGKTTMMKAAREA
WEVGGYRVVGAALAGKAAEGLEKEAGIQSRTLASWELRWKEGRDTLDSSTVLVLDEAGMIASKQMARFVE
AAVKAGAKLVLIGDPDQLQPIEAGAAFRAVVERIGYAELETIYRQREDWKRAASMDLARGRIAEALSAYQ
QQGRVLGSVLKAEAVASLIADWNRDYDPARSSLILAHLRRDVRMLNTMAREKLVERGLLSEGHAFRTEDG
VRRFDVGDQIVFLKNDSALGVKNGMIGKVVEAAPNRIVAAIGEGDQRRQVTVDQHVYRNLDHGYATTIHK
AQGATVDRVKVLASLSLDRHLTYVALTRHREDMALYYGRRSFAFNGGLAKVLSRRNAKETTLDYERGTLY
RQALRFAENRGLHIVNVARTLLRDRVDWTVRQKRKFADLAQRLRGIGARLGLFDPRQTHSPKEAAAMVAG
VTLFTKSIKDTVEEKLRANPAVTKQWDEVSTRFRYVFADPETAFRAMNFDAVLSDPTAAKTALDRLVSEP
QALGPLKGKTGLLAGKADREDRRVAELNVTALKRDIERYLNLREAAVQKLQADEQALRHRVSIDIPALSP
AAHRVLERVRDAIDRNDLPAALGYAISNREVKGEIDGFNKAIAERFGERTLLTNAAREPAGKIFDKAADG
LAPGERQKLAEAWPTMRTAQQLAAQERTAATLKQAETLRQSQRQTPVMKQ
MAITHFTPQLIGRGNGRSAVLSAAYRHCARMEYEAEARTVDYSNKRNLAHEEFLLPADAPEWARALIADR
SVAGAVEAFWNRVEAFEKREDAQFGREFIIALPVELSKEQNIALMRQFVLEQVLARGQVADWVYHDEPGN
PHVHLMTTLRPLTEDGFGPKRIPVIGEDGEVLRNKAGKIVYRLWSGEKTEFLEQRNGWLDLQNQHLALAG
LEIRVDGRSYAERGIDLVPTTHIGVGAKAIQRKSTREGEAVDLERLKLFEAQRTENRRRILLRPEIVLDL
LSSEKSVFDQRDIVKVLHRYVDDAGTFQQLMARILQSPELLRIERESVDFATGERLPARYTTRELIRVEA
GMARQAIWLSGRSSHGVRERVLEQVFARHERLSEEQRTAIEHVTKPGGIAAVVGRAGAGKTTMMKAAREA
WEVGGYRVVGAALAGKAAEGLEKEAGIQSRTLASWELRWKEGRDTLDSSTVLVLDEAGMIASKQMARFVE
AAVKAGAKLVLIGDPDQLQPIEAGAAFRAVVERIGYAELETIYRQREDWKRAASMDLARGRIAEALSAYQ
QQGRVLGSVLKAEAVASLIADWNRDYDPARSSLILAHLRRDVRMLNTMAREKLVERGLLSEGHAFRTEDG
VRRFDVGDQIVFLKNDSALGVKNGMIGKVVEAAPNRIVAAIGEGDQRRQVTVDQHVYRNLDHGYATTIHK
AQGATVDRVKVLASLSLDRHLTYVALTRHREDMALYYGRRSFAFNGGLAKVLSRRNAKETTLDYERGTLY
RQALRFAENRGLHIVNVARTLLRDRVDWTVRQKRKFADLAQRLRGIGARLGLFDPRQTHSPKEAAAMVAG
VTLFTKSIKDTVEEKLRANPAVTKQWDEVSTRFRYVFADPETAFRAMNFDAVLSDPTAAKTALDRLVSEP
QALGPLKGKTGLLAGKADREDRRVAELNVTALKRDIERYLNLREAAVQKLQADEQALRHRVSIDIPALSP
AAHRVLERVRDAIDRNDLPAALGYAISNREVKGEIDGFNKAIAERFGERTLLTNAAREPAGKIFDKAADG
LAPGERQKLAEAWPTMRTAQQLAAQERTAATLKQAETLRQSQRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4490 | GenBank | WP_244451511 |
| Name | traG_K9D25_RS24475_ZM13|pD |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 69041.33 Da Isoelectric Point: 9.7468
>WP_244451511.1 Ti-type conjugative transfer system protein TraG [Ancylobacter polymorphus]
MNRLLIAMIQVVVMVAVAIFTAGAEHWLAGFGKTEAARLMLGRAGLALPYVLSAGVGVILLFAAAGAIAI
RAVGWGVVTGSAAVIGIATIREGARLAALADRVPAGQYVLAYADPGTMIGAAIALVCAMFALRVAIRGNA
AFAAASPRRIKGRRAIHGETDWMRMETAGKLFGDAGGIVIGERYRVDRDSVAGIAFRADSRDSWGSGGRS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPSALKWGGGLVVLDPSSEVAPMVIGHRRKAGRKVVVLDPA
DAATGFNALDWIGRFGGTKEEDIVAVATWVMTDTARQASARDDFFRASAMQLLTALIADVCLSGHTEKKD
QTLRQVRANLSEPEPKLRERLTRIYEQSGSDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYAAL
ISGHSFTTDELADGGTDIFIALDLKVLEAHPGLARVIIGALMNAIYNRNGEVKDRTLFLLDEVARLGFLR
ILETARDAGRKYGITMTLLFQSIGQMREAYGGRDATSKWFESASWISFAAINDPETADYISRRCGDTTVE
VDQLSRSSQMSGASRTRSKQLARRPLILPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMRSCVGEN
RFHRKEAVR
MNRLLIAMIQVVVMVAVAIFTAGAEHWLAGFGKTEAARLMLGRAGLALPYVLSAGVGVILLFAAAGAIAI
RAVGWGVVTGSAAVIGIATIREGARLAALADRVPAGQYVLAYADPGTMIGAAIALVCAMFALRVAIRGNA
AFAAASPRRIKGRRAIHGETDWMRMETAGKLFGDAGGIVIGERYRVDRDSVAGIAFRADSRDSWGSGGRS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPSALKWGGGLVVLDPSSEVAPMVIGHRRKAGRKVVVLDPA
DAATGFNALDWIGRFGGTKEEDIVAVATWVMTDTARQASARDDFFRASAMQLLTALIADVCLSGHTEKKD
QTLRQVRANLSEPEPKLRERLTRIYEQSGSDFVKENVAVFVNMTPETFSGVYANAVKETHWLSYPNYAAL
ISGHSFTTDELADGGTDIFIALDLKVLEAHPGLARVIIGALMNAIYNRNGEVKDRTLFLLDEVARLGFLR
ILETARDAGRKYGITMTLLFQSIGQMREAYGGRDATSKWFESASWISFAAINDPETADYISRRCGDTTVE
VDQLSRSSQMSGASRTRSKQLARRPLILPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMRSCVGEN
RFHRKEAVR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4491 | GenBank | WP_244451470 |
| Name | t4cp2_K9D25_RS24575_ZM13|pD |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91292.72 Da Isoelectric Point: 5.9109
>WP_244451470.1 conjugal transfer protein TrbE [Ancylobacter polymorphus]
MVAIRSFRHSGPSFSDLVPYAAIVANGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
TGWMIQVEAVRVATSDYPAREECHFPDAVTRAIDDERRAHFQEGRGHFESRHALILTWRPPERRRSGLAR
YVYSDSESRAARYADTVLDNFLTSIREVEQYLGNVLSIRRMVTRSVEERGGFQTARYDELFQFIRFCITG
ENHPVRLPEIPMYLDWLATAEFSHGLTPMVDSRYLAVVGIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDALAMVAETEDAIAQASSQLVAYGYYTPVIVL
FDDSEARLREKAEAVRRLVQAEGFGARIETINATEAFLGSLPGNWYANIREPLINTRNLADLIPLNSVWS
GEPTAPCPYYPPGSPPLMLVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYAQAQIFA
FDKGRSMLPLTLAAGGDHYEIGESDGAGAPTLAFCPLAELRTDGDRAWASEWIETLVALQGVTITPDHRN
AISRQIGLMASANGRSLSDFVSGVQMREIKEALHHYTVDGPMGQLLDAESDGLSLRSFQTFEIEQLMNMG
ERSLVPVLLYLFRRIEKRLTGAPSLIVLDEAWLMLGHPTFRDKIREWLKVLRKANCAVILATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDMALGP
VALSFAGASGKEDLKQIAALKAEHGDGWPARWLQQRGIGDAEALLHQS
MVAIRSFRHSGPSFSDLVPYAAIVANGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
TGWMIQVEAVRVATSDYPAREECHFPDAVTRAIDDERRAHFQEGRGHFESRHALILTWRPPERRRSGLAR
YVYSDSESRAARYADTVLDNFLTSIREVEQYLGNVLSIRRMVTRSVEERGGFQTARYDELFQFIRFCITG
ENHPVRLPEIPMYLDWLATAEFSHGLTPMVDSRYLAVVGIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDALAMVAETEDAIAQASSQLVAYGYYTPVIVL
FDDSEARLREKAEAVRRLVQAEGFGARIETINATEAFLGSLPGNWYANIREPLINTRNLADLIPLNSVWS
GEPTAPCPYYPPGSPPLMLVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYAQAQIFA
FDKGRSMLPLTLAAGGDHYEIGESDGAGAPTLAFCPLAELRTDGDRAWASEWIETLVALQGVTITPDHRN
AISRQIGLMASANGRSLSDFVSGVQMREIKEALHHYTVDGPMGQLLDAESDGLSLRSFQTFEIEQLMNMG
ERSLVPVLLYLFRRIEKRLTGAPSLIVLDEAWLMLGHPTFRDKIREWLKVLRKANCAVILATQSISDAER
SGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDMALGP
VALSFAGASGKEDLKQIAALKAEHGDGWPARWLQQRGIGDAEALLHQS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 23981..50200
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| K9D25_RS24440 (K9D25_24440) | 19516..19674 | + | 159 | Protein_18 | DUF736 family protein | - |
| K9D25_RS24445 (K9D25_24445) | 19743..19937 | + | 195 | WP_244451506 | hypothetical protein | - |
| K9D25_RS24450 (K9D25_24450) | 19989..20237 | - | 249 | WP_244451507 | WGR domain-containing protein | - |
| K9D25_RS24455 (K9D25_24455) | 20322..20795 | - | 474 | WP_244451508 | thermonuclease family protein | - |
| K9D25_RS24460 (K9D25_24460) | 21388..21750 | + | 363 | WP_244451509 | hypothetical protein | - |
| K9D25_RS24465 (K9D25_24465) | 22109..22612 | + | 504 | WP_244451510 | hypothetical protein | - |
| K9D25_RS24470 (K9D25_24470) | 22720..23892 | - | 1173 | WP_244451526 | DUF1173 domain-containing protein | - |
| K9D25_RS24475 (K9D25_24475) | 23981..25900 | - | 1920 | WP_244451511 | Ti-type conjugative transfer system protein TraG | virb4 |
| K9D25_RS24480 (K9D25_24480) | 25893..26108 | - | 216 | WP_244451512 | type IV conjugative transfer system coupling protein TraD | - |
| K9D25_RS24485 (K9D25_24485) | 26113..26409 | - | 297 | WP_244451513 | conjugal transfer protein TraC | - |
| K9D25_RS24490 (K9D25_24490) | 26663..29965 | + | 3303 | WP_244451514 | Ti-type conjugative transfer relaxase TraA | - |
| K9D25_RS24495 (K9D25_24495) | 29962..30489 | + | 528 | WP_244451515 | conjugative transfer signal peptidase TraF | - |
| K9D25_RS24500 (K9D25_24500) | 30491..31645 | + | 1155 | WP_244451516 | conjugal transfer protein TraB | - |
| K9D25_RS24505 (K9D25_24505) | 31660..32265 | + | 606 | WP_244451517 | TraH family protein | virB1 |
| K9D25_RS24510 (K9D25_24510) | 32463..34238 | - | 1776 | WP_244451518 | phospholipase D-like domain-containing protein | - |
| K9D25_RS24515 (K9D25_24515) | 34235..37780 | - | 3546 | WP_244451519 | AAA domain-containing protein | - |
| K9D25_RS24520 (K9D25_24520) | 37948..38592 | - | 645 | WP_244451520 | RES family NAD+ phosphorylase | - |
| K9D25_RS24525 (K9D25_24525) | 38589..39386 | - | 798 | WP_244451521 | hypothetical protein | - |
| K9D25_RS24530 (K9D25_24530) | 39512..39787 | - | 276 | WP_244451522 | hypothetical protein | - |
| K9D25_RS24535 (K9D25_24535) | 40164..40484 | + | 321 | WP_244451523 | transcriptional repressor TraM | - |
| K9D25_RS24540 (K9D25_24540) | 40676..41962 | - | 1287 | WP_244451524 | IncP-type conjugal transfer protein TrbI | virB10 |
| K9D25_RS24545 (K9D25_24545) | 41975..42427 | - | 453 | WP_244451466 | conjugal transfer protein TrbH | - |
| K9D25_RS24550 (K9D25_24550) | 42431..43207 | - | 777 | WP_244451527 | P-type conjugative transfer protein TrbG | virB9 |
| K9D25_RS24555 (K9D25_24555) | 43285..43944 | - | 660 | WP_244451467 | conjugal transfer protein TrbF | virB8 |
| K9D25_RS24560 (K9D25_24560) | 43963..45150 | - | 1188 | WP_244451468 | P-type conjugative transfer protein TrbL | virB6 |
| K9D25_RS24565 (K9D25_24565) | 45144..45344 | - | 201 | WP_244451469 | entry exclusion protein TrbK | - |
| K9D25_RS24570 (K9D25_24570) | 45341..46099 | - | 759 | WP_244451528 | P-type conjugative transfer protein TrbJ | virB5 |
| K9D25_RS24575 (K9D25_24575) | 46113..48569 | - | 2457 | WP_244451470 | conjugal transfer protein TrbE | virb4 |
| K9D25_RS24580 (K9D25_24580) | 48579..48878 | - | 300 | WP_029349440 | conjugal transfer protein TrbD | virB3 |
| K9D25_RS24585 (K9D25_24585) | 48871..49242 | - | 372 | WP_126280986 | TrbC/VirB2 family protein | virB2 |
| K9D25_RS24590 (K9D25_24590) | 49232..50200 | - | 969 | WP_029349434 | P-type conjugative transfer ATPase TrbB | virB11 |
| K9D25_RS24595 (K9D25_24595) | 50216..50839 | - | 624 | WP_244451529 | acyl-homoserine-lactone synthase TraI | - |
| K9D25_RS24600 (K9D25_24600) | 51224..52441 | + | 1218 | WP_244451471 | plasmid partitioning protein RepA | - |
| K9D25_RS24605 (K9D25_24605) | 52441..53469 | + | 1029 | WP_244451472 | plasmid partitioning protein RepB | - |
| K9D25_RS24610 (K9D25_24610) | 53744..55093 | + | 1350 | WP_244451473 | plasmid replication protein RepC | - |
Host bacterium
| ID | 7187 | GenBank | NZ_CP083243 |
| Plasmid name | ZM13|pD | Incompatibility group | - |
| Plasmid size | 83314 bp | Coordinate of oriT [Strand] | 26553..26607 [-] |
| Host baterium | Ancylobacter polymorphus strain ZM13 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |