Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106729 |
| Name | oriT_pAtAG67 |
| Organism | Agrobacterium fabrum strain LBA645 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP079858 (57510..57550 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 18..24, 29..35 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pAtAG67
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGTGCTTGC
TCCAAGGGCGCAATTATACGTCGCTGACGCGACGTGCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file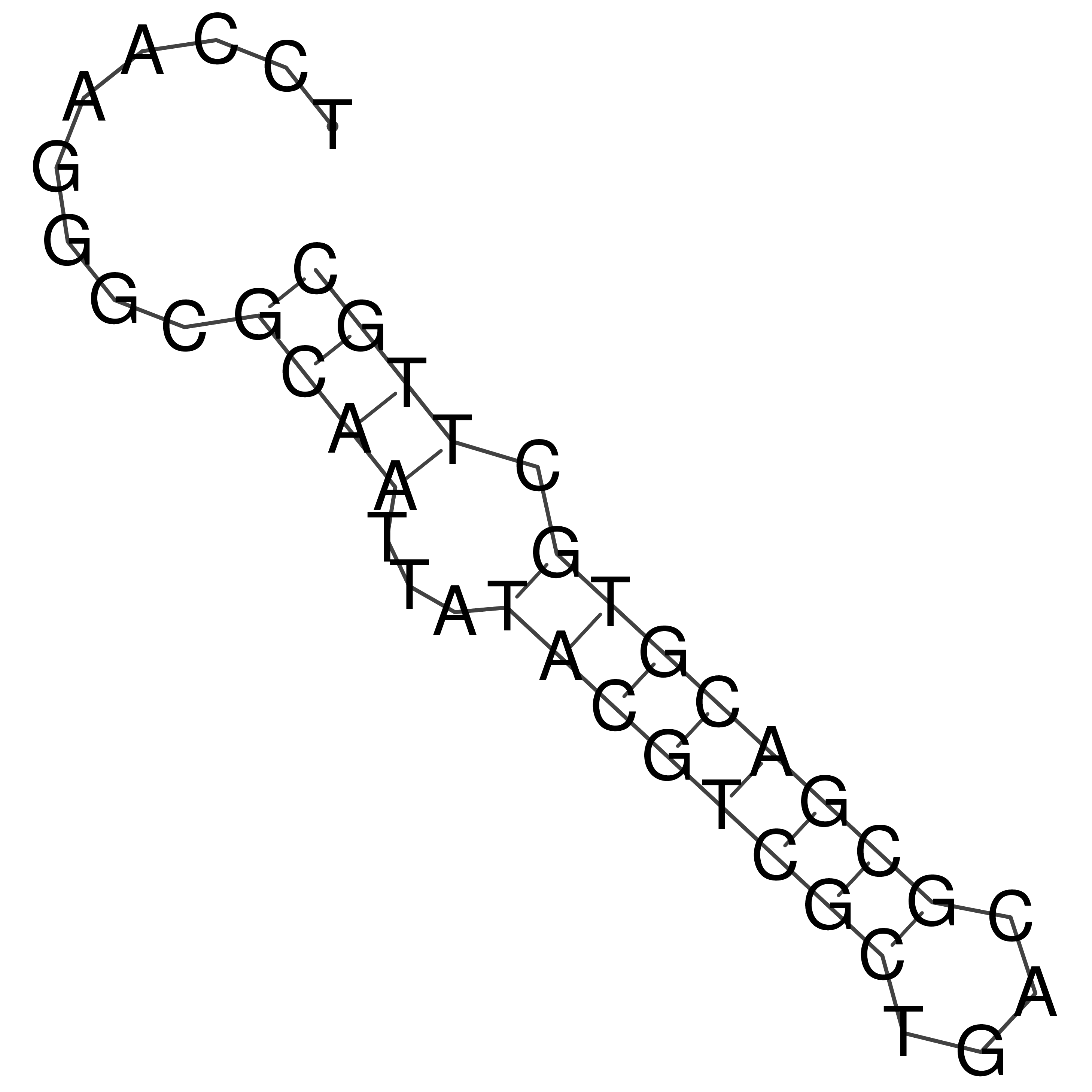
Relaxase
| ID | 4444 | GenBank | WP_243823448 |
| Name | traA_KXJ62_RS25900_pAtAG67 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123609.01 Da Isoelectric Point: 9.7191
>WP_243823448.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium fabrum]
MAIAHFSASIVSRGGGRSAVLSAAYRHCAKMEYEREARAIDYTRKQGLLHEEFVVPADAPKWVRALISDR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNITLVRDFAEKHILAKGMVADWVYHGNPGN
PHIHLMTTLRPLTEDGFGSKKVAVIGEDGQPVRTKSGKILYQLWAGSTDDFNVLRDGWFDRLNHHLALGG
IDLKIDGRSYEKQGIELEPTIHLGVGAKAIERKAQQQGVRPELERIELNEQRRSENTRRILNNPAIVLDL
ITREKSVFDERDVAKVLHRYVDDPTVFQQLMLRIIQNPEVFRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMVRQALWLSNRDGHPVSEAALAATFRRHERLSDEQKTAIERIAGPARIGAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIESRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
AVVRAGARIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANARITGTGLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNIMAREKLVERGIVGEGHVFKTADG
IRQFDVGDQIVFLKNEGSLGVKNGMIAHVVEVAPNRIVAVIGEGDQRRQVFVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAEVLSRKNAKETTLDYERGNLY
REALRFAESRGLNIVQVARTLVRDRLDWTLRQKTRLVDLGQRLAVFAERLGLHHPPKTQTMKEAAPMIAG
IKTFSGSVADTVENKLDTDPALRQQWEEVSTRFSYVFADPETAFRAMNFDAVLADKETAKLALQTLTTAP
ASIGPLKGKTGILASKTEREDRRIAERNVPALKRDLEQYLRMRETAVQRLQADEQALRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERTMLTNAAREPSGKLFDKLSEG
MQPEQKERLKEAWPIMRTSQQLAAHERTVQTLRQAEDLRLAQRQTPVMKQ
MAIAHFSASIVSRGGGRSAVLSAAYRHCAKMEYEREARAIDYTRKQGLLHEEFVVPADAPKWVRALISDR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNITLVRDFAEKHILAKGMVADWVYHGNPGN
PHIHLMTTLRPLTEDGFGSKKVAVIGEDGQPVRTKSGKILYQLWAGSTDDFNVLRDGWFDRLNHHLALGG
IDLKIDGRSYEKQGIELEPTIHLGVGAKAIERKAQQQGVRPELERIELNEQRRSENTRRILNNPAIVLDL
ITREKSVFDERDVAKVLHRYVDDPTVFQQLMLRIIQNPEVFRLQRDTIEFATGEKVPARYSTRAMIRLEA
TMVRQALWLSNRDGHPVSEAALAATFRRHERLSDEQKTAIERIAGPARIGAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIESRTLASWELRWNRGRDVLDDKTVFVMDEAGMVASKQMAGFVD
AVVRAGARIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALTAYD
ANARITGTGLKAEAVERLIGDWNHDYDQTKTTLILAHLRRDVRMLNIMAREKLVERGIVGEGHVFKTADG
IRQFDVGDQIVFLKNEGSLGVKNGMIAHVVEVAPNRIVAVIGEGDQRRQVFVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGRRSFAFNGGLAEVLSRKNAKETTLDYERGNLY
REALRFAESRGLNIVQVARTLVRDRLDWTLRQKTRLVDLGQRLAVFAERLGLHHPPKTQTMKEAAPMIAG
IKTFSGSVADTVENKLDTDPALRQQWEEVSTRFSYVFADPETAFRAMNFDAVLADKETAKLALQTLTTAP
ASIGPLKGKTGILASKTEREDRRIAERNVPALKRDLEQYLRMRETAVQRLQADEQALRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERTMLTNAAREPSGKLFDKLSEG
MQPEQKERLKEAWPIMRTSQQLAAHERTVQTLRQAEDLRLAQRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4476 | GenBank | WP_243823447 |
| Name | traG_KXJ62_RS25885_pAtAG67 |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 70998.41 Da Isoelectric Point: 9.2530
>WP_243823447.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens complex]
MTMNRLLLLILPAMIMLAAMFATSGMEQRLAAFGTSPQAKLMLGRAGLALPYIAAVAIGIIGLFAANGSA
NIKAAGLSVLAGSGVVITIATLSEVIRLNSIASSVPAEQSVLACADPVTMIGASIAFISGMFALRVAIKG
NAAFATTAPKRIGGKRAVHGEADWMKIQEAAKLFPERGGIIIGERYRVDRDSVAAMPFRAVDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVCEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTET
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGNVKGRTLFLLDEVARLGY
LRILETARDAGRKYGIMLTMIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
ENRFHRTGTGTDTHTEHAPPWQKEGTRP
MTMNRLLLLILPAMIMLAAMFATSGMEQRLAAFGTSPQAKLMLGRAGLALPYIAAVAIGIIGLFAANGSA
NIKAAGLSVLAGSGVVITIATLSEVIRLNSIASSVPAEQSVLACADPVTMIGASIAFISGMFALRVAIKG
NAAFATTAPKRIGGKRAVHGEADWMKIQEAAKLFPERGGIIIGERYRVDRDSVAAMPFRAVDRQSWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSSEVAPMVCEHRRQAGRKVIVLD
PTAGGVGFNALDWIGRHGNTKEEDIVAVATWIMTDNPRTASARDDFFRASAMQLLTALIADVCLSGHTET
EDQTLRRVRANLSEPEPKLRARLTKIYEGSESDFVKENVSVFVNMTPETFSGVYANAVKETHWLSYPNYA
GLVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGNVKGRTLFLLDEVARLGY
LRILETARDAGRKYGIMLTMIFQSLGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSTGMKGSSRSRSKQLSRRPLILPHEVLRMRSDEQIVFTSGNPPLRCGRAIWFRRDDMKACVG
ENRFHRTGTGTDTHTEHAPPWQKEGTRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4477 | GenBank | WP_060716498 |
| Name | t4cp2_KXJ62_RS26220_pAtAG67 |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 92090.79 Da Isoelectric Point: 5.4919
>WP_060716498.1 MULTISPECIES: conjugal transfer protein TrbE [Rhizobium/Agrobacterium group]
MVALNLFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINMIMSRLG
SGWMIQVEALRVPTIDYPADDACHFADPVTRAIDTERRTHFERESGHFESRHALILTWRPPEPRRSGLAR
YVYSDTASRSATYADTALESFRTSIREVEQYLANVVSIRRMVTRETEERGGFRIARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVESRFLGVVAIDGLAAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVL
FDEQQERLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLSLIAAQFRRYADAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDAAEGGERDSPRLAFCPLAELASDGDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGAFQCFEVEEL
MNLGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRNKIREWLKVVRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHFEHGADWPKHWLQQRGIAHAETLFQAA
MVALNLFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINMIMSRLG
SGWMIQVEALRVPTIDYPADDACHFADPVTRAIDTERRTHFERESGHFESRHALILTWRPPEPRRSGLAR
YVYSDTASRSATYADTALESFRTSIREVEQYLANVVSIRRMVTRETEERGGFRIARYDELFQFIRFCITG
ENHPVRLPDIPMYLDWLVTAELQHGLTPTVESRFLGVVAIDGLAAESWPGILNSLDLMPLTYRWSSRFVF
LDEQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMMMVAETEDAIAEASSQLVAYGYYTPVIVL
FDEQQERLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLVNTRNLADLIPLNSVWS
GSPVAPCPFYPPGSPPLMQVASGSTPFRLNLHVDDVGHTLVFGPTGSGKSTLLSLIAAQFRRYADAQIFA
FDKGRSMLPLTLAIGGDHYEIGGDAAEGGERDSPRLAFCPLAELASDGDRAWAAEWIETLVALQGVTVTP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEQDGLALGAFQCFEVEEL
MNLGERNLVPVLLYMFRRVEKRLTGAPSLIILDEAWLMLGHPTFRNKIREWLKVVRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATALPKREYYVASPEGRRLFDM
SLGPVALSFVGASGKEDLKRIRALHFEHGADWPKHWLQQRGIAHAETLFQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 121164..130840
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KXJ62_RS26170 (KXJ62_26060) | 116801..117586 | + | 786 | WP_060716474 | amino acid ABC transporter ATP-binding protein | - |
| KXJ62_RS26175 (KXJ62_26065) | 117732..119069 | + | 1338 | WP_243823404 | MmgE/PrpD family protein | - |
| KXJ62_RS26180 (KXJ62_26070) | 119245..121110 | + | 1866 | WP_243823405 | LysR family transcriptional regulator | - |
| KXJ62_RS26185 (KXJ62_26075) | 121164..122474 | - | 1311 | WP_153517050 | IncP-type conjugal transfer protein TrbI | virB10 |
| KXJ62_RS26190 (KXJ62_26080) | 122487..122966 | - | 480 | WP_243823406 | conjugal transfer protein TrbH | - |
| KXJ62_RS26195 (KXJ62_26085) | 122966..123820 | - | 855 | WP_071208371 | P-type conjugative transfer protein TrbG | virB9 |
| KXJ62_RS26200 (KXJ62_26090) | 123838..124500 | - | 663 | WP_153517051 | conjugal transfer protein TrbF | virB8 |
| KXJ62_RS26205 (KXJ62_26095) | 124515..125711 | - | 1197 | WP_243823407 | P-type conjugative transfer protein TrbL | virB6 |
| KXJ62_RS26210 (KXJ62_26100) | 125705..125923 | - | 219 | WP_243823408 | entry exclusion protein TrbK | - |
| KXJ62_RS26215 (KXJ62_26105) | 125920..126729 | - | 810 | WP_153517054 | P-type conjugative transfer protein TrbJ | virB5 |
| KXJ62_RS26220 (KXJ62_26110) | 126701..129169 | - | 2469 | WP_060716498 | conjugal transfer protein TrbE | virb4 |
| KXJ62_RS26225 (KXJ62_26115) | 129180..129479 | - | 300 | WP_045023373 | conjugal transfer protein TrbD | virB3 |
| KXJ62_RS26230 (KXJ62_26120) | 129472..129879 | - | 408 | WP_060716499 | conjugal transfer pilin TrbC | virB2 |
| KXJ62_RS26235 (KXJ62_26125) | 129869..130840 | - | 972 | WP_060716500 | P-type conjugative transfer ATPase TrbB | virB11 |
| KXJ62_RS26240 (KXJ62_26130) | 130837..131472 | - | 636 | WP_243823409 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 7166 | GenBank | NZ_CP079858 |
| Plasmid name | pAtAG67 | Incompatibility group | - |
| Plasmid size | 131838 bp | Coordinate of oriT [Strand] | 57510..57550 [+] |
| Host baterium | Agrobacterium fabrum strain LBA645 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |