Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106622 |
| Name | oriT_pLN1-233332 |
| Organism | Agrobacterium tumefaciens strain LN-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP092894 (89863..89891 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pLN1-233332
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file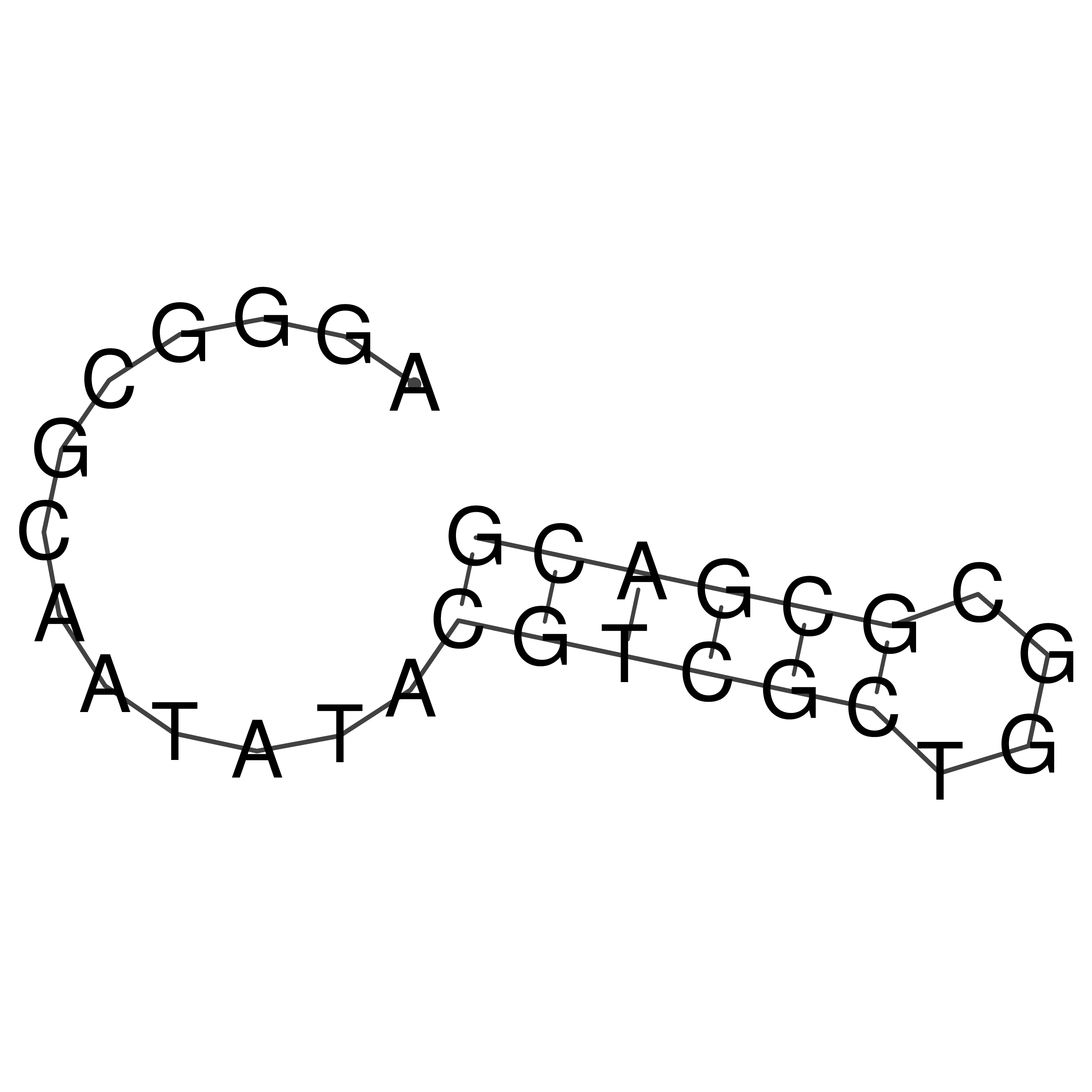
Relaxase
| ID | 4391 | GenBank | WP_243017639 |
| Name | traA_MLE07_RS24930_pLN1-233332 |
UniProt ID | _ |
| Length | 1538 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1538 a.a. Molecular weight: 170632.14 Da Isoelectric Point: 8.3007
>WP_243017639.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGEAELKHEELALPDRVPAWLRTAIDG
KMVAGASEALWNAVDAFETRADAQLARELIIALPEELTRGENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGQKKVPVLGADGEALRVVTPDRPNGKIVYKLWAGDKETMKAWKIAWADTANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLADPALLLKQ
LANERSTFDEKDIATALHRYVDDPSDFVNIRARLMASDDLVLLKPQQVDRQTGKVSEPAVFTTRDILRLE
YDMAQSAEILSQRKGFGIGEGRIAAAVRSVETADPEKSFTLDREQVEAVRHVTGDNGIAAVVGLAGAGKS
TLLAAARVAWESENHRVFGAALAGKAAESLEDSSGIKSRTLASWELAWANGRDLLERGDVFVVDEAGMVS
SQQMARILKTAEDASVKVVLVGDAMQLQPIQAGAAFRAISDRIGSAELAGVRRQREEWARGASRLFARGK
VEEALDAYAREGRIFAAESRADVVERIVADWSGARRDLIRSTANSEQPVRLRGDELLVLAHTNDDVKRLN
EALRKVVKDDGALSDARDFQTERGVREFAAGDRIIFLENARFLEPRARHRGPQYVKNGMLGMVVSTGDNR
GDPLLSVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHMTYVSMTRHRDRADL
YAAREDFQPKPEWGESRVDHAAGVTGELVETGMAKFRPQDEDADESPYADVKTDDGAVHRLWGVSLPKAL
EQVGVSEGDTVALRKDGVERVTVTVPVVDESTGNKSFEERTVDRNVWTAELKETADVRRARIERESHRPD
LFKQLVERLGRSGAKTTTLDFESEAGYQVHAHDFARRRGIDLAAVVAAGMEEGASRRLAWLAEKREQVAK
LWQRASVALGFAIEREKRVSYNEERAQTLSAQVSANRKYLIAPTTMFSRSVDEEARRAQLASARWNERDA
ILRPVLQKIYRDPDGALARLNVLASDVRIEPRKLADDLATKPQRLGRLRGSELLVDGRAARTERDTAMSA
LSELLPLVRAHATEFRRQAERFGIREEQRRAHMSLSVPALSKQAMARLVEIEAVRQRGGQDAYKTAFTYA
VEDRLLVQEVKAVNEALTARFGWSAFTEKADAIAERNMVERMPEDLAPDHREKLARLFAVVRRFAEEQHL
AERKDRSKIVAGASVEPGRETMSVLPILAAVTEFKTPVEDEARERARAVAHYGHNRAELAETATRIWRDP
ASAVARIEALIVKGFAGDRIAAAVANDPAAYGALRGSDRIMDRLLAVGRERKDALLAVPEAESRVRALGP
SYATALDTETRAITEERRRMGVAIPGLSVSAEEALRELSAAMKKKDAKLEAAAGSFDPAIRREFDAVSRA
LDERFGRNAILRGEKDVINRVAPAQRRAFETMQERLKTVQQIVRMQASQEVVAERQRRVLDRARGVIR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGEAELKHEELALPDRVPAWLRTAIDG
KMVAGASEALWNAVDAFETRADAQLARELIIALPEELTRGENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEEGFGQKKVPVLGADGEALRVVTPDRPNGKIVYKLWAGDKETMKAWKIAWADTANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLADPALLLKQ
LANERSTFDEKDIATALHRYVDDPSDFVNIRARLMASDDLVLLKPQQVDRQTGKVSEPAVFTTRDILRLE
YDMAQSAEILSQRKGFGIGEGRIAAAVRSVETADPEKSFTLDREQVEAVRHVTGDNGIAAVVGLAGAGKS
TLLAAARVAWESENHRVFGAALAGKAAESLEDSSGIKSRTLASWELAWANGRDLLERGDVFVVDEAGMVS
SQQMARILKTAEDASVKVVLVGDAMQLQPIQAGAAFRAISDRIGSAELAGVRRQREEWARGASRLFARGK
VEEALDAYAREGRIFAAESRADVVERIVADWSGARRDLIRSTANSEQPVRLRGDELLVLAHTNDDVKRLN
EALRKVVKDDGALSDARDFQTERGVREFAAGDRIIFLENARFLEPRARHRGPQYVKNGMLGMVVSTGDNR
GDPLLSVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHMTYVSMTRHRDRADL
YAAREDFQPKPEWGESRVDHAAGVTGELVETGMAKFRPQDEDADESPYADVKTDDGAVHRLWGVSLPKAL
EQVGVSEGDTVALRKDGVERVTVTVPVVDESTGNKSFEERTVDRNVWTAELKETADVRRARIERESHRPD
LFKQLVERLGRSGAKTTTLDFESEAGYQVHAHDFARRRGIDLAAVVAAGMEEGASRRLAWLAEKREQVAK
LWQRASVALGFAIEREKRVSYNEERAQTLSAQVSANRKYLIAPTTMFSRSVDEEARRAQLASARWNERDA
ILRPVLQKIYRDPDGALARLNVLASDVRIEPRKLADDLATKPQRLGRLRGSELLVDGRAARTERDTAMSA
LSELLPLVRAHATEFRRQAERFGIREEQRRAHMSLSVPALSKQAMARLVEIEAVRQRGGQDAYKTAFTYA
VEDRLLVQEVKAVNEALTARFGWSAFTEKADAIAERNMVERMPEDLAPDHREKLARLFAVVRRFAEEQHL
AERKDRSKIVAGASVEPGRETMSVLPILAAVTEFKTPVEDEARERARAVAHYGHNRAELAETATRIWRDP
ASAVARIEALIVKGFAGDRIAAAVANDPAAYGALRGSDRIMDRLLAVGRERKDALLAVPEAESRVRALGP
SYATALDTETRAITEERRRMGVAIPGLSVSAEEALRELSAAMKKKDAKLEAAAGSFDPAIRREFDAVSRA
LDERFGRNAILRGEKDVINRVAPAQRRAFETMQERLKTVQQIVRMQASQEVVAERQRRVLDRARGVIR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4422 | GenBank | WP_220318755 |
| Name | traG_MLE07_RS24915_pLN1-233332 |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70624.35 Da Isoelectric Point: 9.1364
>WP_220318755.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Agrobacterium]
MTIRAKPHLSLFLILVPITVTAFAAYVVGWHWPELAADMSGKTQYWFLRASPVPVLLFGPLAGLLTVWAL
PLHRRKPIAMATLICFLSVAGFYGLREFGRLSPSVESGAVTWNRALSFIDIVAVVGAVVGFMATAASARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVVGERYRVDREIVHELPFDPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPIVVEHRRENLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQNSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFRASDLIGGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFERRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLLYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDMKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKDMDTQAK
PNRFAKLRP
MTIRAKPHLSLFLILVPITVTAFAAYVVGWHWPELAADMSGKTQYWFLRASPVPVLLFGPLAGLLTVWAL
PLHRRKPIAMATLICFLSVAGFYGLREFGRLSPSVESGAVTWNRALSFIDIVAVVGAVVGFMATAASARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVVGERYRVDREIVHELPFDPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPIVVEHRRENLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQNSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFRASDLIGGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFERRALFMLDEVDLLGYMRVL
EEARDRGRKYGVSMMLLYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDMKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKDMDTQAK
PNRFAKLRP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 55293..65722
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MLE07_RS24710 (MLE07_24710) | 50341..50574 | + | 234 | Protein_49 | transposase | - |
| MLE07_RS24715 (MLE07_24715) | 50805..52166 | + | 1362 | WP_243017635 | IS110 family transposase | - |
| MLE07_RS24720 (MLE07_24720) | 52176..53060 | + | 885 | Protein_51 | IS3 family transposase | - |
| MLE07_RS24725 (MLE07_24725) | 53306..53548 | + | 243 | WP_142173158 | hypothetical protein | - |
| MLE07_RS24730 (MLE07_24730) | 54295..54657 | - | 363 | WP_142173160 | transcriptional regulator | - |
| MLE07_RS24735 (MLE07_24735) | 54751..55290 | + | 540 | WP_223215662 | hypothetical protein | - |
| MLE07_RS24740 (MLE07_24740) | 55293..55979 | + | 687 | WP_220318783 | lytic transglycosylase domain-containing protein | virB1 |
| MLE07_RS24745 (MLE07_24745) | 55976..56275 | + | 300 | WP_220318782 | TrbC/VirB2 family protein | virB2 |
| MLE07_RS24750 (MLE07_24750) | 56277..56618 | + | 342 | WP_220318781 | type IV secretion system protein VirB3 | virB3 |
| MLE07_RS24755 (MLE07_24755) | 56690..57453 | + | 764 | Protein_58 | IS5 family transposase | - |
| MLE07_RS24760 (MLE07_24760) | 57450..59840 | + | 2391 | WP_243017637 | VirB4 family type IV secretion system protein | virb4 |
| MLE07_RS24765 (MLE07_24765) | 59840..60538 | + | 699 | WP_220318779 | P-type DNA transfer protein VirB5 | virB5 |
| MLE07_RS24770 (MLE07_24770) | 60535..60765 | + | 231 | WP_220318778 | EexN family lipoprotein | - |
| MLE07_RS24775 (MLE07_24775) | 60770..61705 | + | 936 | WP_220318777 | type IV secretion system protein | virB6 |
| MLE07_RS24780 (MLE07_24780) | 61741..62007 | + | 267 | WP_223215661 | hypothetical protein | - |
| MLE07_RS24785 (MLE07_24785) | 62011..62682 | + | 672 | WP_220318776 | virB8 family protein | virB8 |
| MLE07_RS24790 (MLE07_24790) | 62682..63530 | + | 849 | WP_220318802 | P-type conjugative transfer protein VirB9 | virB9 |
| MLE07_RS24795 (MLE07_24795) | 63543..64712 | + | 1170 | WP_220318775 | type IV secretion system protein VirB10 | virB10 |
| MLE07_RS24800 (MLE07_24800) | 64721..65722 | + | 1002 | WP_220318774 | P-type DNA transfer ATPase VirB11 | virB11 |
| MLE07_RS24805 (MLE07_24805) | 65778..66122 | - | 345 | WP_220318773 | hypothetical protein | - |
| MLE07_RS24810 (MLE07_24810) | 66574..68733 | + | 2160 | WP_220318772 | ParB/RepB/Spo0J family partition protein | - |
| MLE07_RS24815 (MLE07_24815) | 68801..69457 | + | 657 | WP_220318771 | hypothetical protein | - |
| MLE07_RS24820 (MLE07_24820) | 69656..70077 | + | 422 | Protein_71 | antirestriction protein | - |
Host bacterium
| ID | 7059 | GenBank | NZ_CP092894 |
| Plasmid name | pLN1-233332 | Incompatibility group | - |
| Plasmid size | 233332 bp | Coordinate of oriT [Strand] | 89863..89891 [+] |
| Host baterium | Agrobacterium tumefaciens strain LN-1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsH, arsC |
| Degradation gene | - |
| Symbiosis gene | fixB, fixA |
| Anti-CRISPR | - |