Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106321 |
| Name | oriT_pRK1-5 |
| Organism | Cupriavidus basilensis strain DSM 11853 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP062809 (15825..15921 [+], 97 nt) |
| oriT length | 97 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 97 nt
GAATAAGGGACAGCGAAGATAGATAACCGGCCCGCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGTTGATAACACCAAGGAAAGTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file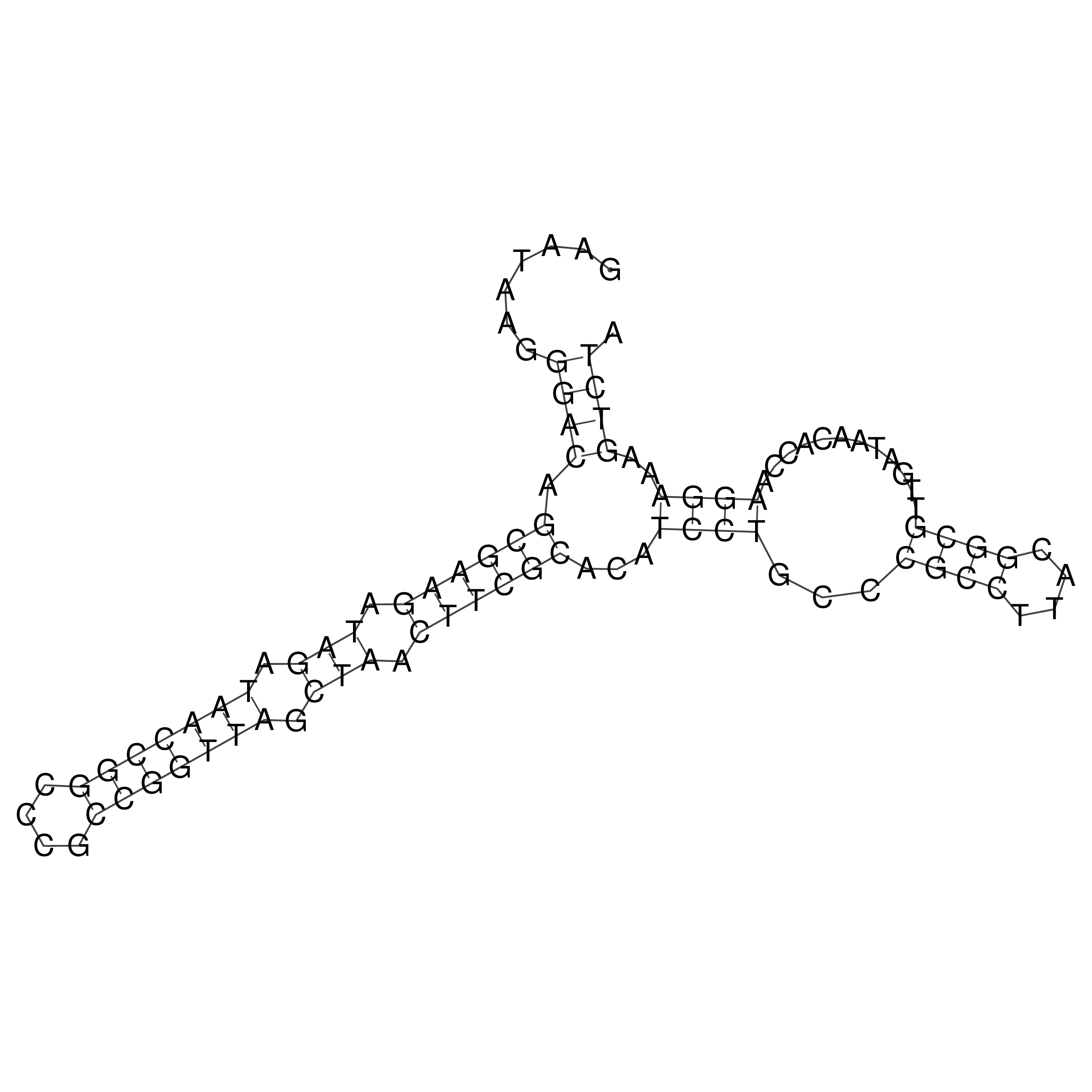
Relaxase
| ID | 4228 | GenBank | WP_012478238 |
| Name | Relaxase_F7R26_RS40385_pRK1-5 |
UniProt ID | A0A1H7CDK8 |
| Length | 752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 752 a.a. Molecular weight: 82867.55 Da Isoelectric Point: 10.6815
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTALERDYGLERDNHEPRQRGAEGRAADMEQHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GLELRERANGLVIEAGDGTMVKASTLARDLSKPKLEARLGPFEASPERQTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRKDRLVEAAKRSGRLRRATIKVIGESGPNKKLLYAQASKALRDE
IQAINKQYQQDRQALYDSHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDPRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMERYGSRITVNGTTQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTERADQRPAEHRGRAGRGPGGPGQRAATHDDAARPIAVA
RAGFGRPGGGRAGTAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRDVPRHL
EQQGTQPDHQLRRDISRPGSGVAAAPPGLAAADKYIAERESKRSKGFDIPKHARYTAGEGGLAFEGVRNI
EGQALALLKRGDEVMVMPIDQATARRMSRIAVGDAVTITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1H7CDK8 |
Auxiliary protein
| ID | 1883 | GenBank | WP_011255188 |
| Name | WP_011255188_pRK1-5 |
UniProt ID | A0A1H7CD62 |
| Length | 123 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 123 a.a. Molecular weight: 13964.97 Da Isoelectric Point: 8.4531
MEADEQPTAKRRQHLRVPVFPDEKEQIEANAKRAGVSVARYLRDVGQGYQIRGVMDYEYVRELVRVNGDL
GRLGGLLKLWLTDDPRTARFGDATILALLGRIEATQDEMSRLMKSVVQPRAEP
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1H7CD62 |
T4CP
| ID | 4248 | GenBank | WP_012478237 |
| Name | t4cp2_F7R26_RS40380_pRK1-5 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69616.60 Da Isoelectric Point: 8.4953
MKIKMNNAVGPQVRSAKPKPSKLLPILGGVSLVGGLQAATQFFAYTFQYHASLGANLGHVYAPWSILNWS
AKWYSQYPDEIMRAGSIGMVTATVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKDAPTATGVYVGGWQDKDGNFYYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASSSGGVCWNPLDEIRVGTEAEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKARNDGGTATLPSVDAMLADPNRDIGELWMEMTTYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARSVSRSDFRIKDLMHSDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFENGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLRSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNDKGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFQARASIPAPKATDRLRQVVEAGE
GITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4249 | GenBank | WP_011013264 |
| Name | t4cp2_F7R26_RS40520_pRK1-5 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 94125.12 Da Isoelectric Point: 6.3997
MIEAIAIGIAVLGAVLLLILFARIRQVDAELKLKKHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YQGDDNASSTEEQREMVSFRINQALAGLGSGWMVHVDAVRRPAPNYSERGVSTFTDPVSAAIDEERRRLF
EGLGTMYEGHFVFAVTWFPPVLAQRKFVELMFDDDAVTPDHTARTVGLIEQFKREIRSLESRMSSAVKLT
RLKGHKIATEEGREVTHDDFLSWLQFCVTGNYHPVQLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVS
LEGFPLESTPGLLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSVNQD
AATMVADAEAAIAEVNSGLVAAGYFTSVVVLMDEDRERLAAAALSVEKAINRLAFAARIETINTMDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLAPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAAINAGSKGKTGLHFTVAADDDRLAFC
PLQFLETKADRAWAMEWIDTILALNGVDTTPGQRNEIGHAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERGLKGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTAALYRRMG
LNKRQIEILATAVPKRQYYYVSENGRRLYDLALGPLALAFVGASDKESVAAIKSLQAKFGHEWVDHWLAG
RNLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 30549..41172
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| F7R26_RS40475 (F7R26_040465) | 25637..26065 | - | 429 | WP_011114069 | antirestriction protein | - |
| F7R26_RS40480 (F7R26_040470) | 26238..26501 | + | 264 | WP_011114070 | type II toxin-antitoxin system RelB/DinJ family antitoxin | - |
| F7R26_RS40485 (F7R26_040475) | 26488..26769 | + | 282 | WP_011114071 | type II toxin-antitoxin system YafQ family toxin | - |
| F7R26_RS40490 (F7R26_040480) | 28155..29378 | - | 1224 | WP_011013257 | plasmid replication initiator TrfA | - |
| F7R26_RS40495 (F7R26_040485) | 29427..29765 | - | 339 | WP_011013259 | single-stranded DNA-binding protein | - |
| F7R26_RS40500 (F7R26_040490) | 29875..30234 | + | 360 | WP_011013260 | transcriptional regulator | - |
| F7R26_RS40505 (F7R26_040495) | 30549..31514 | + | 966 | WP_011013261 | P-type conjugative transfer ATPase TrbB | virB11 |
| F7R26_RS40510 (F7R26_040500) | 31544..32008 | + | 465 | WP_011013262 | conjugal transfer system pilin TrbC | virB2 |
| F7R26_RS40515 (F7R26_040505) | 32012..32323 | + | 312 | WP_011013263 | conjugal transfer protein TrbD | virB3 |
| F7R26_RS40520 (F7R26_040510) | 32320..34878 | + | 2559 | WP_011013264 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| F7R26_RS40525 (F7R26_040515) | 34875..35663 | + | 789 | WP_011013265 | conjugal transfer protein TrbF | virB8 |
| F7R26_RS40530 (F7R26_040520) | 35681..36574 | + | 894 | WP_014386236 | P-type conjugative transfer protein TrbG | virB9 |
| F7R26_RS40535 (F7R26_040525) | 36577..37038 | + | 462 | WP_011013267 | lipoprotein | - |
| F7R26_RS40540 (F7R26_040530) | 37042..38421 | + | 1380 | WP_012478192 | TrbI/VirB10 family protein | virB10 |
| F7R26_RS40545 (F7R26_040535) | 38438..39211 | + | 774 | WP_011255135 | P-type conjugative transfer protein TrbJ | virB5 |
| F7R26_RS40550 (F7R26_040540) | 39221..39442 | + | 222 | WP_011255136 | entry exclusion lipoprotein TrbK | - |
| F7R26_RS40555 (F7R26_040545) | 39454..41172 | + | 1719 | WP_012478193 | P-type conjugative transfer protein TrbL | virB6 |
| F7R26_RS40560 (F7R26_040550) | 41191..41769 | + | 579 | WP_011255138 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| F7R26_RS40565 (F7R26_040555) | 41771..42406 | + | 636 | WP_011255139 | transglycosylase SLT domain-containing protein | - |
| F7R26_RS40570 (F7R26_040560) | 42436..42705 | + | 270 | WP_011255140 | hypothetical protein | - |
| F7R26_RS40575 (F7R26_040565) | 42702..43400 | + | 699 | WP_012478194 | TraX family protein | - |
| F7R26_RS40580 (F7R26_040570) | 43416..43859 | + | 444 | WP_011255142 | OmpA family protein | - |
| F7R26_RS40585 (F7R26_040575) | 43962..44378 | + | 417 | WP_012478195 | hypothetical protein | - |
Host bacterium
| ID | 6758 | GenBank | NZ_CP062809 |
| Plasmid name | pRK1-5 | Incompatibility group | - |
| Plasmid size | 81787 bp | Coordinate of oriT [Strand] | 15825..15921 [+] |
| Host baterium | Cupriavidus basilensis strain DSM 11853 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |