Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106320 |
| Name | oriT_XtLr8|unnamed1 |
| Organism | Xanthomonas translucens pv. undulosa strain XtLr8 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP063994 (3507..3604 [+], 98 nt) |
| oriT length | 98 nt |
| IRs (inverted repeats) | 24..30, 36..42 (TAACCGG..CCGGTTA) |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 98 nt
>oriT_XtLr8|unnamed1
GAATAAGGGACAGCGAAGATAGATAACCGGCTCGCCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGGTGATAACGCCAAGGAAAGTCTA
GAATAAGGGACAGCGAAGATAGATAACCGGCTCGCCCGGTTAGCTAACTTCGCACATCCTGCCCGCCTTACGGCGGTGATAACGCCAAGGAAAGTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file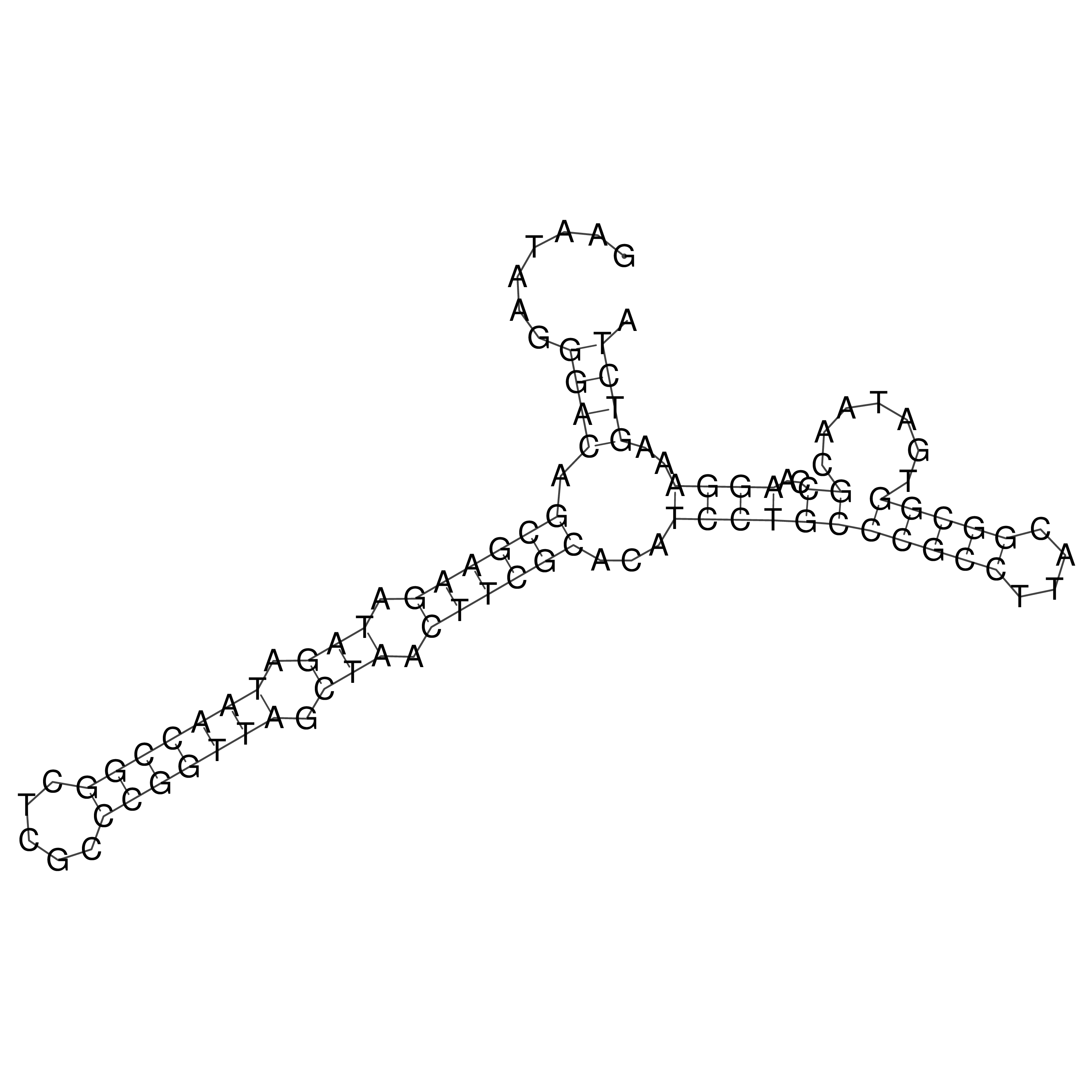
Relaxase
| ID | 4227 | GenBank | WP_058358167 |
| Name | Relaxase_ISN38_RS19540_XtLr8|unnamed1 |
UniProt ID | _ |
| Length | 757 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 757 a.a. Molecular weight: 83436.27 Da Isoelectric Point: 10.8897
>WP_058358167.1 TraI/MobA(P) family conjugative relaxase [Xanthomonas translucens]
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTTLERDYGLERDNHEPRQRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRAN
GLELRERANGLVIEAGDGTMLKASTLARDLSKPKLEARLGPFEASPERHTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRRDRLVEAAKRSGRLRRATIKVIGENGANKKLLYAQASNALRDE
IQAINKQYQQDRQALYESHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDSRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMGRYGSRITVNGTKQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTEQADQRPAEHRGRAGRGAGGPGQRTAAHDDAARPVAIA
RAGIGRPGRGRPGRGRADAAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRD
VPRHLEQQGTQPDHQLRRGISRPGTGVAAAPPGLTAADKYIAEREAKRSKGFDIPKHARYTAGDGALAFQ
GVRNIEGQALALLKRGDEVMVMPIDQATARRLSRIAVSEAVTITPKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLVNYVTDAQSKDHRLGQVQITNCDAVSVQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICAGLGYGEHQRVSAVHNDTDNLHIHIAINKIHPTRNTMHEPYYSHRA
LAELCTTLERDYGLERDNHEPRQRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRAN
GLELRERANGLVIEAGDGTMLKASTLARDLSKPKLEARLGPFEASPERHTRTKAKRQYQKDPIRLRVNTV
ELYARYKAEQQTLTATRGQALEQARRRRDRLVEAAKRSGRLRRATIKVIGENGANKKLLYAQASNALRDE
IQAINKQYQQDRQALYESHSRRTWADWLKKEATHGNAEALTALRAREAAQGLKGNTIQGRGDSRPGHAPV
TDNITKKGTIIFRAGLTAVRDDGDKLQVSREATREGLQEALRLAMGRYGSRITVNGTKQFQAQIIRAAVD
SQLPITFADPALESRRLALLKKENTHERPDRTEQADQRPAEHRGRAGRGAGGPGQRTAAHDDAARPVAIA
RAGIGRPGRGRPGRGRADAAGVHAGAVHRKPDIGRIGRVPPPQSQHRLRTLSQLGVVRIAGGSEVLLPRD
VPRHLEQQGTQPDHQLRRGISRPGTGVAAAPPGLTAADKYIAEREAKRSKGFDIPKHARYTAGDGALAFQ
GVRNIEGQALALLKRGDEVMVMPIDQATARRLSRIAVSEAVTITPKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4247 | GenBank | WP_206712257 |
| Name | t4cp2_ISN38_RS19670_XtLr8|unnamed1 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93919.14 Da Isoelectric Point: 6.4347
>WP_206712257.1 VirB4 family type IV secretion/conjugal transfer ATPase [Xanthomonas translucens]
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSLAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLITQFKREITSLDSRLSSAVKLT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVVGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFATNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKTVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGSATAPCPMYPPLAPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGAMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNIYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVAAIKSLEAKFGHEWLHEWLAG
RGLNLNDYLEAA
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSLAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLITQFKREITSLDSRLSSAVKLT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVVGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFATNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKTVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSTIWTGSATAPCPMYPPLAPALMHCVTHGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGAMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPAVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNIYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVAAIKSLEAKFGHEWLHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 17415..28173
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ISN38_RS19625 (ISN38_19615) | 12899..13117 | - | 219 | WP_015913624 | hypothetical protein | - |
| ISN38_RS19630 (ISN38_19620) | 13163..13591 | - | 429 | WP_058358154 | antirestriction protein | - |
| ISN38_RS20545 | 13709..13837 | + | 129 | Protein_20 | type II toxin-antitoxin system mRNA interferase toxin, RelE/StbE family | - |
| ISN38_RS19640 (ISN38_19630) | 15152..16375 | - | 1224 | WP_206712255 | plasmid replication initiator TrfA | - |
| ISN38_RS19645 (ISN38_19635) | 16423..16761 | - | 339 | WP_058358152 | single-stranded DNA-binding protein | - |
| ISN38_RS19650 (ISN38_19640) | 16870..17229 | + | 360 | WP_058358151 | transcriptional regulator | - |
| ISN38_RS19655 (ISN38_19645) | 17415..18377 | + | 963 | WP_206712256 | P-type conjugative transfer ATPase TrbB | virB11 |
| ISN38_RS19660 (ISN38_19650) | 18394..18825 | + | 432 | WP_058358149 | IncP-type conjugal transfer pilin TrbC | virB2 |
| ISN38_RS19665 (ISN38_19655) | 18829..19140 | + | 312 | WP_046833105 | conjugal transfer protein TrbD | virB3 |
| ISN38_RS19670 (ISN38_19660) | 19137..21695 | + | 2559 | WP_206712257 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| ISN38_RS19675 (ISN38_19665) | 21698..22480 | + | 783 | WP_058358172 | conjugal transfer protein TrbF | virB8 |
| ISN38_RS19680 (ISN38_19670) | 22498..23397 | + | 900 | WP_206712258 | P-type conjugative transfer protein TrbG | virB9 |
| ISN38_RS19685 (ISN38_19675) | 23400..23867 | + | 468 | WP_206712238 | conjugal transfer protein TrbH | - |
| ISN38_RS19690 (ISN38_19680) | 23872..25290 | + | 1419 | WP_206712239 | TrbI/VirB10 family protein | virB10 |
| ISN38_RS19695 (ISN38_19685) | 25308..26072 | + | 765 | WP_058358143 | P-type conjugative transfer protein TrbJ | virB5 |
| ISN38_RS19700 (ISN38_19690) | 26082..26309 | + | 228 | WP_058358142 | entry exclusion lipoprotein TrbK | - |
| ISN38_RS19705 (ISN38_19695) | 26320..28173 | + | 1854 | WP_206712240 | P-type conjugative transfer protein TrbL | virB6 |
| ISN38_RS19710 (ISN38_19700) | 28199..28783 | + | 585 | WP_206712241 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| ISN38_RS19715 (ISN38_19705) | 28797..29453 | + | 657 | WP_206712242 | transglycosylase SLT domain-containing protein | - |
| ISN38_RS19720 (ISN38_19710) | 29530..30165 | + | 636 | WP_241754010 | TraX family protein | - |
| ISN38_RS19725 (ISN38_19715) | 30181..30501 | + | 321 | WP_206712243 | hypothetical protein | - |
| ISN38_RS19730 (ISN38_19720) | 30521..31792 | + | 1272 | WP_206712244 | hypothetical protein | - |
| ISN38_RS19735 (ISN38_19725) | 31905..32267 | + | 363 | WP_241754008 | hypothetical protein | - |
| ISN38_RS19740 (ISN38_19730) | 32300..32773 | - | 474 | WP_206712245 | hypothetical protein | - |
| ISN38_RS19745 (ISN38_19735) | 32868..33128 | - | 261 | WP_206712246 | hypothetical protein | - |
Host bacterium
| ID | 6757 | GenBank | NZ_CP063994 |
| Plasmid name | XtLr8|unnamed1 | Incompatibility group | IncP1 |
| Plasmid size | 45351 bp | Coordinate of oriT [Strand] | 3507..3604 [+] |
| Host baterium | Xanthomonas translucens pv. undulosa strain XtLr8 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF9 |