Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106309 |
| Name | oriT_1,231,410|unnamed |
| Organism | Enterococcus faecium 1231410 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_GG692535 (39677..39868 [-], 192 nt) |
| oriT length | 192 nt |
| IRs (inverted repeats) | 106..112, 122..128 (ATTTTTT..AAAAAAT) 107..113, 120..126 (TTTTTTG..CAAAAAA) 93..98, 105..110 (AAAATA..TATTTT) 65..71, 75..81 (AGTTGGC..GCCAACT) 41..49, 52..60 (CACCTTCCT..AGGAAGGTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 192 nt
>oriT_1,231,410|unnamed
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCTAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATAGCTTCATATTTTTTGCTATCACAAAAAAATCCATTTTCGACCTATTTTCGGTCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
CCAAGGAATTAATACAAAGAGCATAAGGGAAAACTGATAGCACCTTCCTAAAGGAAGGTGGCTAAGTTGGCTGTGCCAACTGGTTTTCTTTCAAAATAGCTTCATATTTTTTGCTATCACAAAAAAATCCATTTTCGACCTATTTTCGGTCATAATATAGTACCTACTTTTGGTCATAGTTTCGTCCGTAGT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file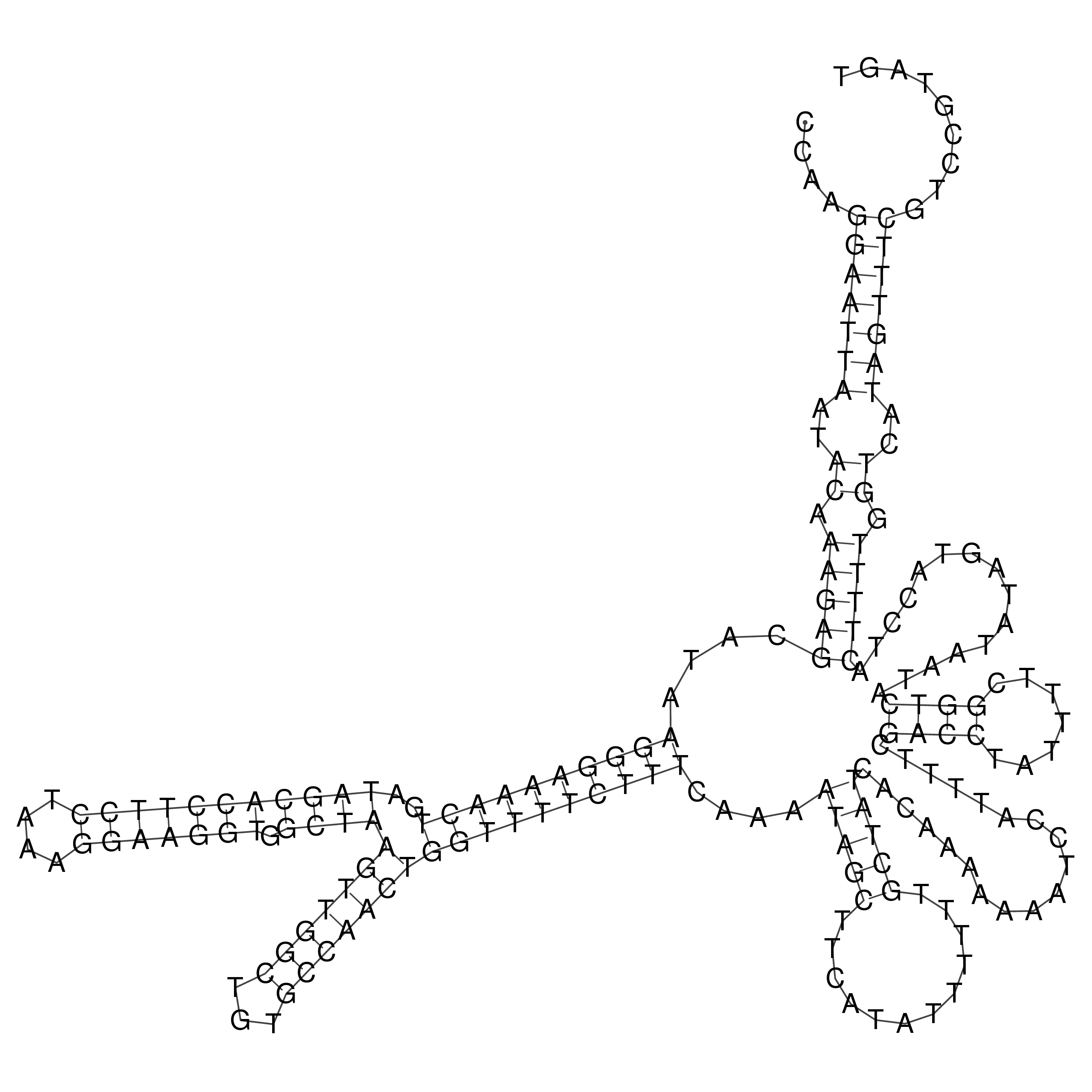
Relaxase
| ID | 4219 | GenBank | WP_002326870 |
| Name | mobP2_EFTG_RS20645_1,231,410|unnamed |
UniProt ID | B5U8V6 |
| Length | 506 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 506 a.a. Molecular weight: 60597.01 Da Isoelectric Point: 9.3485
>WP_002326870.1 MULTISPECIES: MobP2 family relaxase [Enterococcus]
MTEHIFNRKSASIILKAKFETGKSKKNNHMVELQKFVDYISRQEAIRQDKLDHDYSEDELNELARIEKAL
EKIEQETDQPVIKDLREMDKYIDYMTRKKAILENVDNEIVNGAFSNTKRYITKKDISKIKESVIEAKNNG
SVMFQDVISFDNDFLVREGYYNPETNELNENVLYEATKSMMGKMQEKEELVDPFWFATIHRNTEHIHIHV
TAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFGSKILDRTNEFEKISKLRKEVPLELKQSVKDSLI
QLYVENNSRYDKELVKYLKELKKEIPSTTRGYNELPEETKEKIDEVTNYMTKDNPKRKEYDRMTKEIDEL
YQTTYGKRYNPQEYYLNRKKDFNARMGNAVVGQIKLLKQSEEKNLKSNELKNLLDGKKLNVKFDREKPEF
KTKYAKQSYDDFQKRLEERQLAREKNEVHFKKISEKFKDKKLIRKIDRVINDDLNMYRAKNDYEVAQRMI
EQEKMRQAREFEIGMN
MTEHIFNRKSASIILKAKFETGKSKKNNHMVELQKFVDYISRQEAIRQDKLDHDYSEDELNELARIEKAL
EKIEQETDQPVIKDLREMDKYIDYMTRKKAILENVDNEIVNGAFSNTKRYITKKDISKIKESVIEAKNNG
SVMFQDVISFDNDFLVREGYYNPETNELNENVLYEATKSMMGKMQEKEELVDPFWFATIHRNTEHIHIHV
TAMERKNTREIMEYDGVLQARGKRKQSTLDDMIFKFGSKILDRTNEFEKISKLRKEVPLELKQSVKDSLI
QLYVENNSRYDKELVKYLKELKKEIPSTTRGYNELPEETKEKIDEVTNYMTKDNPKRKEYDRMTKEIDEL
YQTTYGKRYNPQEYYLNRKKDFNARMGNAVVGQIKLLKQSEEKNLKSNELKNLLDGKKLNVKFDREKPEF
KTKYAKQSYDDFQKRLEERQLAREKNEVHFKKISEKFKDKKLIRKIDRVINDDLNMYRAKNDYEVAQRMI
EQEKMRQAREFEIGMN
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B5U8V6 |
T4CP
| ID | 4241 | GenBank | WP_002311855 |
| Name | t4cp2_EFTG_RS20585_1,231,410|unnamed |
UniProt ID | _ |
| Length | 952 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 952 a.a. Molecular weight: 108225.12 Da Isoelectric Point: 7.4751
>WP_002311855.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Enterococcus]
MKLVQNNEVKEIKNGFVKNSNKILGKLKKVGSFKKNKYHTGKIEGEDLPKSLQDKLNVPLVFIGVFLGSI
VFLAITYLLNFLKALLVAIGQSESGLLGVEFDPKFSLAYLKPLGDTKQYFMCAVVAVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLEKRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMGTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRKENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGKNYSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYARFIKDLKTRLEYSIINNIPISEEDYQDYLNLSGSQQTINEEELLKLVNTEVKSQDMEGQVLS
EQEMKINIYLKESQKEAIELVKVINGRLADENSADMVIKTINSQILDTPFIQSLINDSIAIHNVQESKDQ
IMILEQQLANAYELAVFKTKNSILKNKIIKLKNAMTKLAKAI
MKLVQNNEVKEIKNGFVKNSNKILGKLKKVGSFKKNKYHTGKIEGEDLPKSLQDKLNVPLVFIGVFLGSI
VFLAITYLLNFLKALLVAIGQSESGLLGVEFDPKFSLAYLKPLGDTKQYFMCAVVAVIGVAYLIYGKLNH
ESEKNLVYGQKGDSRLSTIKEIKRDYHEIPEKTEVYDGIGGVPISHYQDKYYIDRDTVNTVVLGASRSGK
GETFVVPLLDNLSRAKIQSNMVVNDPKGELFSASKETLEKRGYRVLVLNIDDPLESMSFNPLQLVIDSWA
NKDYHEASKRANTLTSMLFASGMGTDNEFFYKSAKSAVNAIILTIVEHCFNNDCIEKITMYNVAQMLNEL
GSLFYTDPKTGKEKNALDEYFNTLPQGNQAKIQYGSTSFAGDKAKGSILSTASQGIEMFTSDLFGKLTSK
MSIDLKEIGFPKSIQFKVNTNLVGKRISISFLRKENDVIRLIKKYRVKVKALGLCVLNFNEYLQDGDMLD
IRYEEEKSRAWYRIEFPKKQANSHDTSLVTKKFKSGKNYSDLEISTLRLKYTDQPTAVFMVIPDYDPSNH
VLASIFISQLYTELASNCKQTPDKKCFRRVHFLLDEFGNMPAIDNMDGIMTVCLGRNMLFDLVIQSYSQL
ETRYDKAFKTIKENCQNHILIMSNDEETIEEISKKCGHKTTINRSASSKHLDTDSSVTSSAEQERVITVE
RASQLIEGEQIILRNLHRQDTKRNKIRPFPIFNTKETNMPYRYQFLAEDFDTQRDINEIDIACEHINLSL
QDNQIPYARFIKDLKTRLEYSIINNIPISEEDYQDYLNLSGSQQTINEEELLKLVNTEVKSQDMEGQVLS
EQEMKINIYLKESQKEAIELVKVINGRLADENSADMVIKTINSQILDTPFIQSLINDSIAIHNVQESKDQ
IMILEQQLANAYELAVFKTKNSILKNKIIKLKNAMTKLAKAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 20848..48348
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EFTG_RS20530 (EFTG_05419) | 16129..19758 | + | 3630 | WP_002311844 | SpaA isopeptide-forming pilin-related protein | - |
| EFTG_RS20535 (EFTG_05420) | 19818..20180 | + | 363 | WP_002311845 | hypothetical protein | - |
| EFTG_RS22270 (EFTG_05421) | 20194..20346 | + | 153 | WP_002311846 | hypothetical protein | - |
| EFTG_RS20540 (EFTG_05422) | 20414..20836 | + | 423 | WP_002311847 | hypothetical protein | - |
| EFTG_RS20545 (EFTG_05423) | 20848..22263 | + | 1416 | WP_002311848 | CpaF/VirB11 family protein | virB11 |
| EFTG_RS20550 (EFTG_05424) | 22280..23095 | + | 816 | WP_002313315 | hypothetical protein | - |
| EFTG_RS20555 (EFTG_05425) | 23095..23892 | + | 798 | WP_002311850 | hypothetical protein | - |
| EFTG_RS20560 (EFTG_05426) | 23910..24272 | + | 363 | WP_002311851 | DUF4320 family protein | - |
| EFTG_RS22515 | 24536..24664 | + | 129 | WP_002317412 | hypothetical protein | - |
| EFTG_RS20570 (EFTG_05427) | 24683..24958 | + | 276 | WP_002311852 | TrbC/VirB2 family protein | virB2 |
| EFTG_RS20575 (EFTG_05428) | 25313..26893 | + | 1581 | WP_224997648 | hypothetical protein | - |
| EFTG_RS20580 (EFTG_05429) | 26906..27622 | + | 717 | WP_002311854 | hypothetical protein | - |
| EFTG_RS20585 (EFTG_05430) | 27635..30493 | + | 2859 | WP_002311855 | VirD4-like conjugal transfer protein, CD1115 family | - |
| EFTG_RS20590 (EFTG_05431) | 30544..33180 | + | 2637 | WP_002313318 | pLS20_p028 family conjugation system transmembrane protein | - |
| EFTG_RS20595 (EFTG_05432) | 33228..33539 | + | 312 | WP_002317413 | DUF5592 family protein | - |
| EFTG_RS20600 (EFTG_05433) | 33536..34153 | + | 618 | WP_002311858 | hypothetical protein | virb4 |
| EFTG_RS20605 (EFTG_05434) | 34169..34636 | + | 468 | WP_002311859 | hypothetical protein | - |
| EFTG_RS20610 (EFTG_05435) | 34652..36607 | + | 1956 | WP_002311860 | tra protein | virb4 |
| EFTG_RS20615 (EFTG_05436) | 36629..37765 | + | 1137 | WP_002311861 | bifunctional lytic transglycosylase/C40 family peptidase | orf14 |
| EFTG_RS20620 (EFTG_05437) | 37780..38430 | + | 651 | WP_002311862 | cell division protein FtsL | - |
| EFTG_RS20625 (EFTG_05438) | 38444..39340 | + | 897 | WP_002311863 | hypothetical protein | - |
| EFTG_RS20630 (EFTG_05439) | 39376..39606 | + | 231 | WP_002311864 | hypothetical protein | - |
| EFTG_RS20635 (EFTG_05440) | 39957..40253 | + | 297 | WP_002311865 | hypothetical protein | - |
| EFTG_RS20640 (EFTG_05441) | 40250..40717 | + | 468 | WP_002311867 | hypothetical protein | - |
| EFTG_RS20645 (EFTG_05442) | 40799..42319 | + | 1521 | WP_002326870 | MobP2 family relaxase | - |
| EFTG_RS20650 (EFTG_05443) | 42341..42670 | + | 330 | WP_002311869 | hypothetical protein | - |
| EFTG_RS20655 (EFTG_05444) | 42672..42935 | + | 264 | WP_002311870 | hypothetical protein | - |
| EFTG_RS20660 (EFTG_05445) | 43110..43976 | + | 867 | WP_002311871 | hypothetical protein | - |
| EFTG_RS20665 (EFTG_05446) | 44131..45957 | + | 1827 | WP_002311872 | ArdC-like ssDNA-binding domain-containing protein | - |
| EFTG_RS20670 (EFTG_05447) | 45965..47038 | + | 1074 | WP_002311873 | hypothetical protein | - |
| EFTG_RS20675 (EFTG_05448) | 47047..47307 | + | 261 | WP_002311874 | hypothetical protein | - |
| EFTG_RS20680 (EFTG_05449) | 47344..48348 | + | 1005 | WP_002311875 | DUF3991 domain-containing protein | traP |
| EFTG_RS20685 (EFTG_05450) | 48373..48729 | + | 357 | WP_002311876 | hypothetical protein | - |
| EFTG_RS20690 (EFTG_05451) | 48891..49082 | + | 192 | WP_002311877 | hypothetical protein | - |
| EFTG_RS20695 (EFTG_05452) | 49191..49409 | + | 219 | WP_002311878 | hypothetical protein | - |
| EFTG_RS20700 (EFTG_05453) | 49446..49763 | + | 318 | WP_002311879 | hypothetical protein | - |
| EFTG_RS20705 (EFTG_05454) | 49774..49971 | + | 198 | WP_002311880 | hypothetical protein | - |
| EFTG_RS20710 (EFTG_05455) | 49968..50936 | + | 969 | WP_002311881 | zeta toxin family protein | - |
| EFTG_RS20715 (EFTG_05456) | 51180..52352 | + | 1173 | WP_002311882 | hypothetical protein | - |
| EFTG_RS20720 (EFTG_05457) | 52912..53289 | - | 378 | WP_002305129 | type II toxin-antitoxin system HicB family antitoxin | - |
Host bacterium
| ID | 6746 | GenBank | NZ_GG692535 |
| Plasmid name | 1,231,410|unnamed | Incompatibility group | - |
| Plasmid size | 60477 bp | Coordinate of oriT [Strand] | 39677..39868 [-] |
| Host baterium | Enterococcus faecium 1231410 |
Cargo genes
| Drug resistance gene | dfrG |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA1 |