Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106306 |
| Name | oriT_EbN1|2 |
| Organism | Aromatoleum aromaticum EbN1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_006824 (58472..58570 [+], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 61..62 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_EbN1|2
GAATAAGGGTTAGCGAAGGTAGATAACCGGCTCCGCTGGTTAGCTAACTTCGCCCATCCTGCCCGCCTTTCGGCGTTCATTACCCCAGGGAAAGTCTAC
GAATAAGGGTTAGCGAAGGTAGATAACCGGCTCCGCTGGTTAGCTAACTTCGCCCATCCTGCCCGCCTTTCGGCGTTCATTACCCCAGGGAAAGTCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file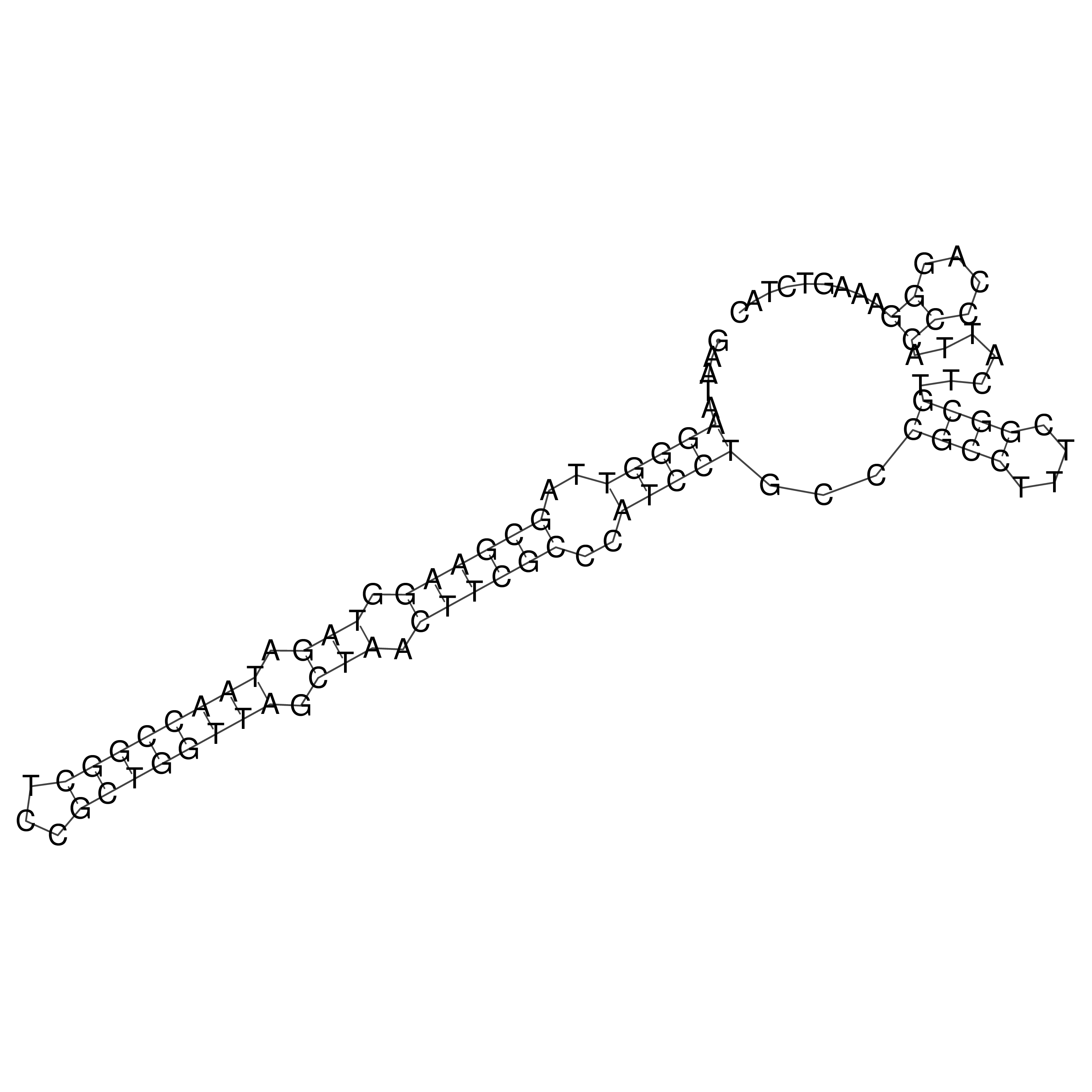
Relaxase
| ID | 4217 | GenBank | WP_011255046 |
| Name | Relaxase_EBN1_RS22090_EbN1|2 |
UniProt ID | Q5NWI9 |
| Length | 740 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 740 a.a. Molecular weight: 81425.10 Da Isoelectric Point: 10.7479
>WP_011255046.1 TraI/MobA(P) family conjugative relaxase [Aromatoleum aromaticum]
MIAKHVPMRSLAKSDFAELVEYITDEQGKTERLGPVTATHCAAATLQAVIGEVLATQRQNTRGAGDKTYH
LLVSFRPGEHPSADTLKVIEERICAGLGYGEHQRVSAVHHDTDNLHLHIAINKIHPERHTMHEPFQSYRT
LAELCAVLERDCGLERDNHQSRRSVAEGRAADMERHAGVESLVGWVRRESLEEMRGAQSWTELHQVMRKN
GLELRERGNGLVVEAADGTRVKASTIARELSKPGLEARLGPFEPSPDRLAEPKMRRQYEKPPIGLRVNTV
ELYARYKAEQQTLTAARADSLAKARRRKDRAVDGAKRANRLRRATITVVDSKGVGKKALYSQASTALRDQ
LAEIHKEYAKEREKLSQGFKRRAWADWLKQEALQGNADALAALRGREAAQGLKGNTIAGQGATKSERAHV
QDNITKKGTIIFRIGQSAVRDDGDKLQVSRDATREGVQAALRLAMERYGSQITVNGTPAFKAQIIRAAVD
SQLPITFADPALELRRQQLLSKEKAHERAARTERGRTDRRRLGGAGSSAAADDNRARTGAGAGRGGSDGR
GGAGMDAPAHVGRKPNVGRIGRVPPPQSQHRLRGMSELSVVRIVGGGEVLLPRDVPRHVEQQRAQSDHAL
RRDAVGVGRLKPAQQAAADEYLAEREDKPLKYFDIPKNSLYNKNARGFSFAGLRHVEGRTLALLRRDDAD
AVMVLPIDQATARRLSRVAVGDPVTVTPRGSIQTSKGRSR
MIAKHVPMRSLAKSDFAELVEYITDEQGKTERLGPVTATHCAAATLQAVIGEVLATQRQNTRGAGDKTYH
LLVSFRPGEHPSADTLKVIEERICAGLGYGEHQRVSAVHHDTDNLHLHIAINKIHPERHTMHEPFQSYRT
LAELCAVLERDCGLERDNHQSRRSVAEGRAADMERHAGVESLVGWVRRESLEEMRGAQSWTELHQVMRKN
GLELRERGNGLVVEAADGTRVKASTIARELSKPGLEARLGPFEPSPDRLAEPKMRRQYEKPPIGLRVNTV
ELYARYKAEQQTLTAARADSLAKARRRKDRAVDGAKRANRLRRATITVVDSKGVGKKALYSQASTALRDQ
LAEIHKEYAKEREKLSQGFKRRAWADWLKQEALQGNADALAALRGREAAQGLKGNTIAGQGATKSERAHV
QDNITKKGTIIFRIGQSAVRDDGDKLQVSRDATREGVQAALRLAMERYGSQITVNGTPAFKAQIIRAAVD
SQLPITFADPALELRRQQLLSKEKAHERAARTERGRTDRRRLGGAGSSAAADDNRARTGAGAGRGGSDGR
GGAGMDAPAHVGRKPNVGRIGRVPPPQSQHRLRGMSELSVVRIVGGGEVLLPRDVPRHVEQQRAQSDHAL
RRDAVGVGRLKPAQQAAADEYLAEREDKPLKYFDIPKNSLYNKNARGFSFAGLRHVEGRTLALLRRDDAD
AVMVLPIDQATARRLSRVAVGDPVTVTPRGSIQTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q5NWI9 |
T4CP
| ID | 4238 | GenBank | WP_011255080 |
| Name | t4cp2_EBN1_RS21890_EbN1|2 |
UniProt ID | _ |
| Length | 845 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 845 a.a. Molecular weight: 93764.76 Da Isoelectric Point: 6.2164
>WP_011255080.1 VirB4 family type IV secretion/conjugal transfer ATPase [Aromatoleum aromaticum]
MIETIAITIAGVGTVLLLILFVRIRQVDAELQLKKHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YRGDDNASSTDEQREMVSFRLNQALAGLGSGWMVHVDAVRRPAPNYSAKGSSHFPDAVSAAIDKERRQLF
EGLGTMYEGYFVLTLTWFPPLLAQRKFVELMFDDDAIAPDRQTRTMGLIEQFKRELTAIENRLSSAVKLM
RMRGQKVVNEDGSTVTHDDFLSWLQFCLTGRHHPILLPSNPVYLDAVIGGQELWGGVVPRIGRNFVQVVA
IEGFPLESTPGLLTALGELPCEYRWSSRFIFMDPHEAVKHLDKFRKKWRQKVRGFFDQVFNTHTGPVDQD
ALSMVGDAEAAIAEVNSGLVAVGYFTGVVILMDENRDKLEESARLVEKSINRLGFAARIETINTLDAFLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGHADAPCPLYPPLSPALMHCVTHGATPFRLNLHVRDIGH
TFMFGPTGAGKSTHLALIAAQLRRYAGMTLYAFDKGMSLYPLTKAVGGLHFSVAADDDRLAFCPLQFLDT
KGDRAWAMEWIDTILALNGVETTPAQRNEIGHAIMSMHRSGGRTLSELSVTIQDEAIREAIRQYTVDGTM
GHLLDAEEDGLSLADFTTFEIEELMDLGEKYALPVLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVLRKANCLVLMATQSLSDVANSGILDVIVESTATKIFLPNVYACDQDTAALYRRMGLNARQIE
ILATAIPKRQYYYVSESGRRLYDLALGPFALAFVGSSDKESVASIKSLEAKFGDQWVHEWLAGKGLNLND
YLEAA
MIETIAITIAGVGTVLLLILFVRIRQVDAELQLKKHRSKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YRGDDNASSTDEQREMVSFRLNQALAGLGSGWMVHVDAVRRPAPNYSAKGSSHFPDAVSAAIDKERRQLF
EGLGTMYEGYFVLTLTWFPPLLAQRKFVELMFDDDAIAPDRQTRTMGLIEQFKRELTAIENRLSSAVKLM
RMRGQKVVNEDGSTVTHDDFLSWLQFCLTGRHHPILLPSNPVYLDAVIGGQELWGGVVPRIGRNFVQVVA
IEGFPLESTPGLLTALGELPCEYRWSSRFIFMDPHEAVKHLDKFRKKWRQKVRGFFDQVFNTHTGPVDQD
ALSMVGDAEAAIAEVNSGLVAVGYFTGVVILMDENRDKLEESARLVEKSINRLGFAARIETINTLDAFLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGHADAPCPLYPPLSPALMHCVTHGATPFRLNLHVRDIGH
TFMFGPTGAGKSTHLALIAAQLRRYAGMTLYAFDKGMSLYPLTKAVGGLHFSVAADDDRLAFCPLQFLDT
KGDRAWAMEWIDTILALNGVETTPAQRNEIGHAIMSMHRSGGRTLSELSVTIQDEAIREAIRQYTVDGTM
GHLLDAEEDGLSLADFTTFEIEELMDLGEKYALPVLLYLFRRIERSLKGQPAVIILDEAWLMLGHPAFRA
KIREWLKVLRKANCLVLMATQSLSDVANSGILDVIVESTATKIFLPNVYACDQDTAALYRRMGLNARQIE
ILATAIPKRQYYYVSESGRRLYDLALGPFALAFVGSSDKESVASIKSLEAKFGDQWVHEWLAGKGLNLND
YLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4239 | GenBank | WP_011255047 |
| Name | t4cp2_EBN1_RS22085_EbN1|2 |
UniProt ID | _ |
| Length | 634 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 634 a.a. Molecular weight: 69736.77 Da Isoelectric Point: 7.6457
>WP_011255047.1 type IV secretory system conjugative DNA transfer family protein [Aromatoleum aromaticum]
MKIKMDNAVGPQVRTKKGAGGKLLPVLGAVSLGAGLQSATQFFAHIFDYQANLGGHFNHVYAPWSILEWS
SKWYSHYPNEIMQAGSVGMMISTVGLLGVAVAKVVSSNSSKANEYLHGSARWAGKKDIQAAGLLPRPRRL
LDVVTGKDTAPSTGVYVGGWEDKDGNFHYLRHNGPEHILTYAPTRSGKGVGLVVPTLLSWGDSAVVTDLK
GELWAMTAGWRKKHAKNNVLRFEPATANGSVCWNPLDEIRMGTEHEVGDVQNLVTLIVDPDGKGLGSHWQ
KTAFALLVGVILHALYKAKDDDTTATLPSVDAMLADPNRDVGELWMEMATYGHVAGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSFLALYRDPVVARNVSRSEFCIRDLMHSDDPVTLYIVTHPNDKDRLRPLVRVMVN
MIVRLLADKMDFDNGRLVAHYKHRLLMMLDEFPSLGKLQILQESLAFVAGYGIKCYLICQDINQLKSRET
GYGPDETITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPRKNADGNIEEPGDMVVYVAGYPAIYGKQPLYFKDPIFLARAAVPAPKDSDRLVQFAEAGE
RITI
MKIKMDNAVGPQVRTKKGAGGKLLPVLGAVSLGAGLQSATQFFAHIFDYQANLGGHFNHVYAPWSILEWS
SKWYSHYPNEIMQAGSVGMMISTVGLLGVAVAKVVSSNSSKANEYLHGSARWAGKKDIQAAGLLPRPRRL
LDVVTGKDTAPSTGVYVGGWEDKDGNFHYLRHNGPEHILTYAPTRSGKGVGLVVPTLLSWGDSAVVTDLK
GELWAMTAGWRKKHAKNNVLRFEPATANGSVCWNPLDEIRMGTEHEVGDVQNLVTLIVDPDGKGLGSHWQ
KTAFALLVGVILHALYKAKDDDTTATLPSVDAMLADPNRDVGELWMEMATYGHVAGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSFLALYRDPVVARNVSRSEFCIRDLMHSDDPVTLYIVTHPNDKDRLRPLVRVMVN
MIVRLLADKMDFDNGRLVAHYKHRLLMMLDEFPSLGKLQILQESLAFVAGYGIKCYLICQDINQLKSRET
GYGPDETITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTFQEVQRPLLT
PDECLRMPGPRKNADGNIEEPGDMVVYVAGYPAIYGKQPLYFKDPIFLARAAVPAPKDSDRLVQFAEAGE
RITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 12707..23216
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EBN1_RS21835 (EB_RS21435) | 8539..9594 | + | 1056 | WP_011255087 | 2-oxoacid:ferredoxin oxidoreductase subunit beta | - |
| EBN1_RS21840 (EB_RS21440) | 9664..10023 | + | 360 | Protein_7 | electron transfer flavoprotein subunit beta/FixA family protein | - |
| EBN1_RS21845 (EB_RS25080) | 10025..10211 | - | 187 | Protein_8 | electron transfer flavoprotein subunit beta/FixA family protein | - |
| EBN1_RS21850 (EB_RS24560) | 10211..10351 | + | 141 | WP_197531916 | hypothetical protein | - |
| EBN1_RS21855 (EB_RS21445) | 10689..10910 | + | 222 | WP_041647956 | hypothetical protein | - |
| EBN1_RS21860 (EB_RS24565) | 11115..11493 | + | 379 | Protein_11 | IS21 family transposase | - |
| EBN1_RS21865 | 11537..11779 | - | 243 | Protein_12 | plasmid replication initiator TrfA | - |
| EBN1_RS21870 (EB_RS21455) | 12049..12411 | + | 363 | WP_011255084 | transcriptional regulator | - |
| EBN1_RS21875 (EB_RS21460) | 12707..13666 | + | 960 | WP_011255083 | P-type conjugative transfer ATPase TrbB | virB11 |
| EBN1_RS21880 (EB_RS21465) | 13680..14105 | + | 426 | WP_011255082 | conjugal transfer system pilin TrbC | virB2 |
| EBN1_RS21885 (EB_RS21470) | 14108..14419 | + | 312 | WP_011255081 | conjugal transfer protein TrbD | virB3 |
| EBN1_RS21890 (EB_RS21475) | 14416..16953 | + | 2538 | WP_011255080 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| EBN1_RS21895 (EB_RS21480) | 16950..17735 | + | 786 | WP_011255079 | conjugal transfer protein TrbF | virB8 |
| EBN1_RS21900 (EB_RS21485) | 17753..18649 | + | 897 | WP_011255078 | P-type conjugative transfer protein TrbG | virB9 |
| EBN1_RS21905 (EB_RS21490) | 18652..19143 | + | 492 | WP_041647958 | conjugal transfer protein TrbH | - |
| EBN1_RS21910 (EB_RS21495) | 19149..20537 | + | 1389 | WP_011255076 | TrbI/VirB10 family protein | virB10 |
| EBN1_RS21915 (EB_RS21500) | 20556..21314 | + | 759 | WP_011255075 | P-type conjugative transfer protein TrbJ | virB5 |
| EBN1_RS21920 (EB_RS21505) | 21329..21559 | + | 231 | WP_011255074 | entry exclusion lipoprotein TrbK | - |
| EBN1_RS21925 (EB_RS21510) | 21570..23216 | + | 1647 | WP_011255073 | P-type conjugative transfer protein TrbL | virB6 |
| EBN1_RS21930 (EB_RS21515) | 23234..23812 | + | 579 | WP_011255072 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| EBN1_RS21935 (EB_RS21520) | 23818..24414 | + | 597 | WP_041648023 | muramidase | - |
| EBN1_RS21940 (EB_RS21525) | 24411..25109 | + | 699 | WP_011255070 | TraX family protein | - |
| EBN1_RS21945 (EB_RS21530) | 25204..25629 | + | 426 | WP_041647959 | hypothetical protein | - |
| EBN1_RS21950 (EB_RS21535) | 25716..26015 | + | 300 | WP_041647960 | DUF4258 domain-containing protein | - |
| EBN1_RS21955 (EB_RS21540) | 26017..27840 | + | 1824 | WP_157866790 | N-6 DNA methylase | - |
Host bacterium
| ID | 6743 | GenBank | NC_006824 |
| Plasmid name | EbN1|2 | Incompatibility group | - |
| Plasmid size | 223670 bp | Coordinate of oriT [Strand] | 58472..58570 [+] |
| Host baterium | Aromatoleum aromaticum EbN1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |