Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106304 |
| Name | oriT_DS5|unnamed |
| Organism | Enterococcus faecalis DS5 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_GG692896 (22414..22463 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_DS5|unnamed
AATATCGCAACATGCTAGCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
AATATCGCAACATGCTAGCATGTTGCTCTGCTCGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file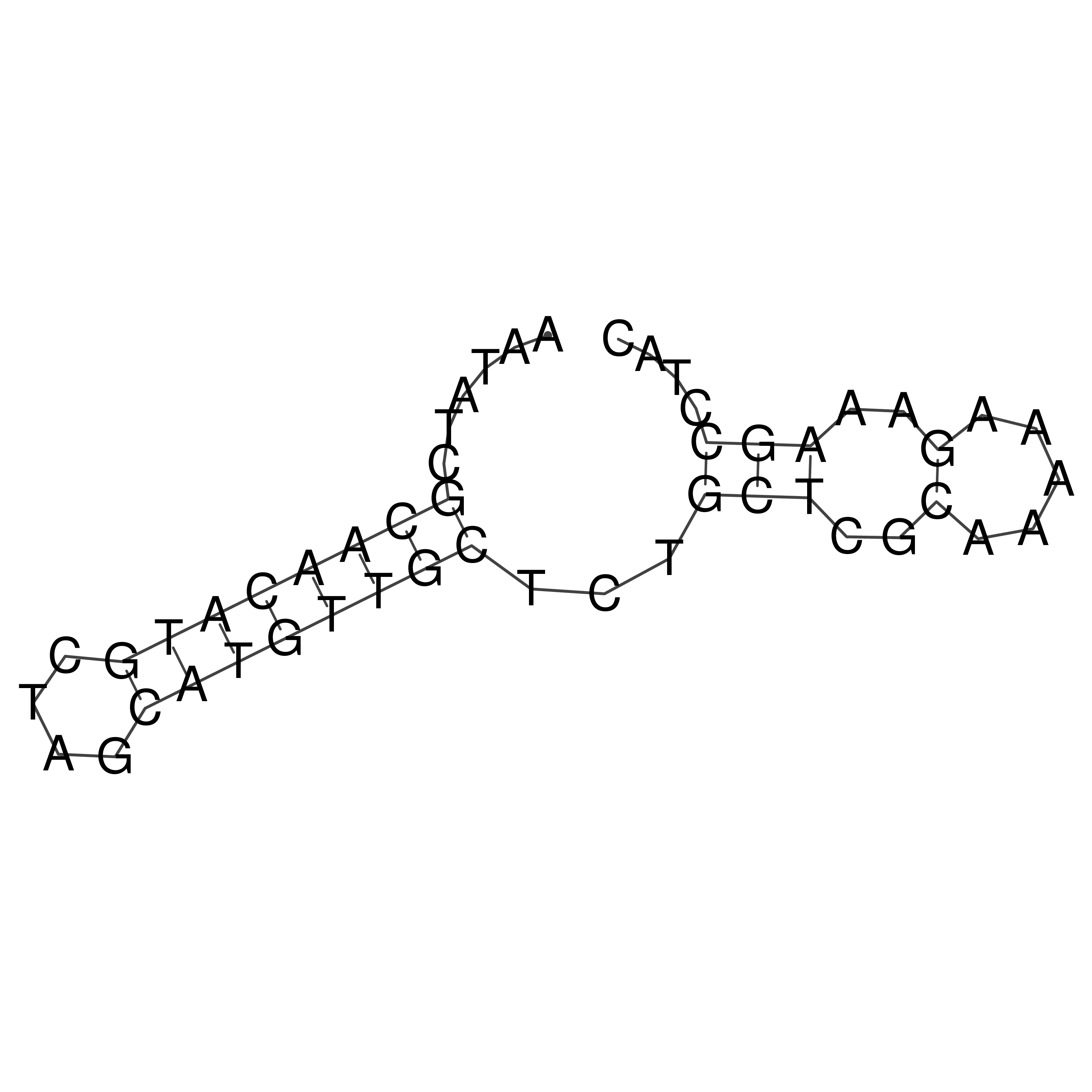
Relaxase
| ID | 4215 | GenBank | WP_002365873 |
| Name | Relaxase_EFEG_RS21310_DS5|unnamed |
UniProt ID | _ |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65469.22 Da Isoelectric Point: 6.0174
>WP_002365873.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKNDPTRYNYEKIVERLKENKSVLTLKEVVEKYVEKSEKTKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
RTIDELVNAINFLAANEIEDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKRVFEQISDKIAAKEGAKIIEKSPKNS
HKKYTMWETESLYKEKIKSRLDFLLEQSSSINDFLEKAEALNLSVDFSKKWTTYRLLDEPQIKNTRSRSL
SKNDPTRYNYEKIVERLKENKSVLTLKEVVEKYVEKSEKTKNEFDYQLTIDEWQISHKTTRGYYVNVDFG
FGERGKVFIGAYKVDPLENGQYNIYVKRKDFFYLMNEKNADRNRYMTGETLIKQLRLYNGQTPLKKEPVM
RTIDELVNAINFLAANEIEDTRQLELLEEKLEAAFLEAEETLETLDEKMLELHQLSNLLLENELNGELEL
IQQKLKTLLPDATLAEFSYEDVRGEIEAIKTSQSLLENKLERTRNEINQLHEIQAVQKKETANQNNVKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1879 | GenBank | WP_002365872 |
| Name | WP_002365872_DS5|unnamed |
UniProt ID | _ |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 13988.05 Da Isoelectric Point: 9.6548
>WP_002365872.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Enterococcus]
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVHSLNALVQSELNKLIKRKEQS
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVHSLNALVQSELNKLIKRKEQS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4237 | GenBank | WP_002365870 |
| Name | t4cp2_EFEG_RS21290_DS5|unnamed |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70184.31 Da Isoelectric Point: 6.5169
>WP_002365870.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNVAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEVKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRALEERKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNVAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEVKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRALEERKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 3466..19769
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EFEG_RS21210 (EFEG_05301) | 1..1412 | + | 1412 | Protein_0 | surface exclusion protein SEA1/PrgA | - |
| EFEG_RS21215 (EFEG_05302) | 3466..5805 | + | 2340 | WP_002365851 | LPXTG-anchored aggregation substance | prgB |
| EFEG_RS21220 (EFEG_05304) | 6442..6798 | + | 357 | WP_002365854 | pheromone response system RNA-binding regulator PrgU | - |
| EFEG_RS21225 (EFEG_05305) | 6826..7701 | + | 876 | WP_002365855 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| EFEG_RS21230 (EFEG_05306) | 7743..8642 | + | 900 | WP_025186515 | hypothetical protein | - |
| EFEG_RS21235 (EFEG_05307) | 8662..9096 | + | 435 | WP_002365857 | hypothetical protein | - |
| EFEG_RS21240 (EFEG_05308) | 9143..9370 | + | 228 | WP_002365858 | hypothetical protein | - |
| EFEG_RS21245 (EFEG_05309) | 9384..9680 | + | 297 | WP_002365860 | hypothetical protein | - |
| EFEG_RS21250 (EFEG_05310) | 9695..10495 | + | 801 | WP_002365861 | PrgH | prgHb |
| EFEG_RS21255 (EFEG_05311) | 10497..10850 | + | 354 | WP_002365862 | PrgI family mobile element protein | prgIc |
| EFEG_RS21260 (EFEG_05312) | 10804..13194 | + | 2391 | WP_002365863 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| EFEG_RS21265 (EFEG_05313) | 13206..15821 | + | 2616 | WP_002365864 | phage tail tip lysozyme | prgK |
| EFEG_RS21270 (EFEG_05314) | 15845..16471 | + | 627 | WP_002365865 | hypothetical protein | prgL |
| EFEG_RS21275 (EFEG_05315) | 16458..16703 | + | 246 | WP_002365866 | hypothetical protein | - |
| EFEG_RS21280 (EFEG_05316) | 16687..17295 | + | 609 | WP_002365867 | hypothetical protein | - |
| EFEG_RS21285 (EFEG_05317) | 17455..17940 | + | 486 | WP_016616031 | PcfB family protein | gbs1365 |
| EFEG_RS21290 (EFEG_05318) | 17940..19769 | + | 1830 | WP_002365870 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| EFEG_RS21295 (EFEG_05319) | 19820..21979 | + | 2160 | WP_002365871 | ArdC-like ssDNA-binding domain-containing protein | - |
| EFEG_RS21300 (EFEG_05320) | 22012..22284 | + | 273 | WP_002362409 | LtrD | - |
| EFEG_RS21305 (EFEG_05321) | 22530..22886 | + | 357 | WP_002365872 | plasmid mobilization relaxosome protein MobC | - |
| EFEG_RS21310 (EFEG_05322) | 22887..24572 | + | 1686 | WP_002365873 | relaxase/mobilization nuclease domain-containing protein | - |
Host bacterium
| ID | 6741 | GenBank | NZ_GG692896 |
| Plasmid name | DS5|unnamed | Incompatibility group | - |
| Plasmid size | 55755 bp | Coordinate of oriT [Strand] | 22414..22463 [+] |
| Host baterium | Enterococcus faecalis DS5 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |