Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106240 |
| Name | oriT_pKP037-3 |
| Organism | Klebsiella pneumoniae strain KP037 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP089873 (7459..7559 [+], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 32..38, 43..49 (TTACGAT..ATCGTAA) 21..26, 38..43 (TTTTCA..TGAAAA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_pKP037-3
AATTAGAATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATAACATACCACGCCTTTTTTAAGG
AATTAGAATAATTTGTTTTGTTTTCAAGCATTTACGATGAAAATCGTAATTGCGTATGGTGTATAGCCGTTAAGGGATAACATACCACGCCTTTTTTAAGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file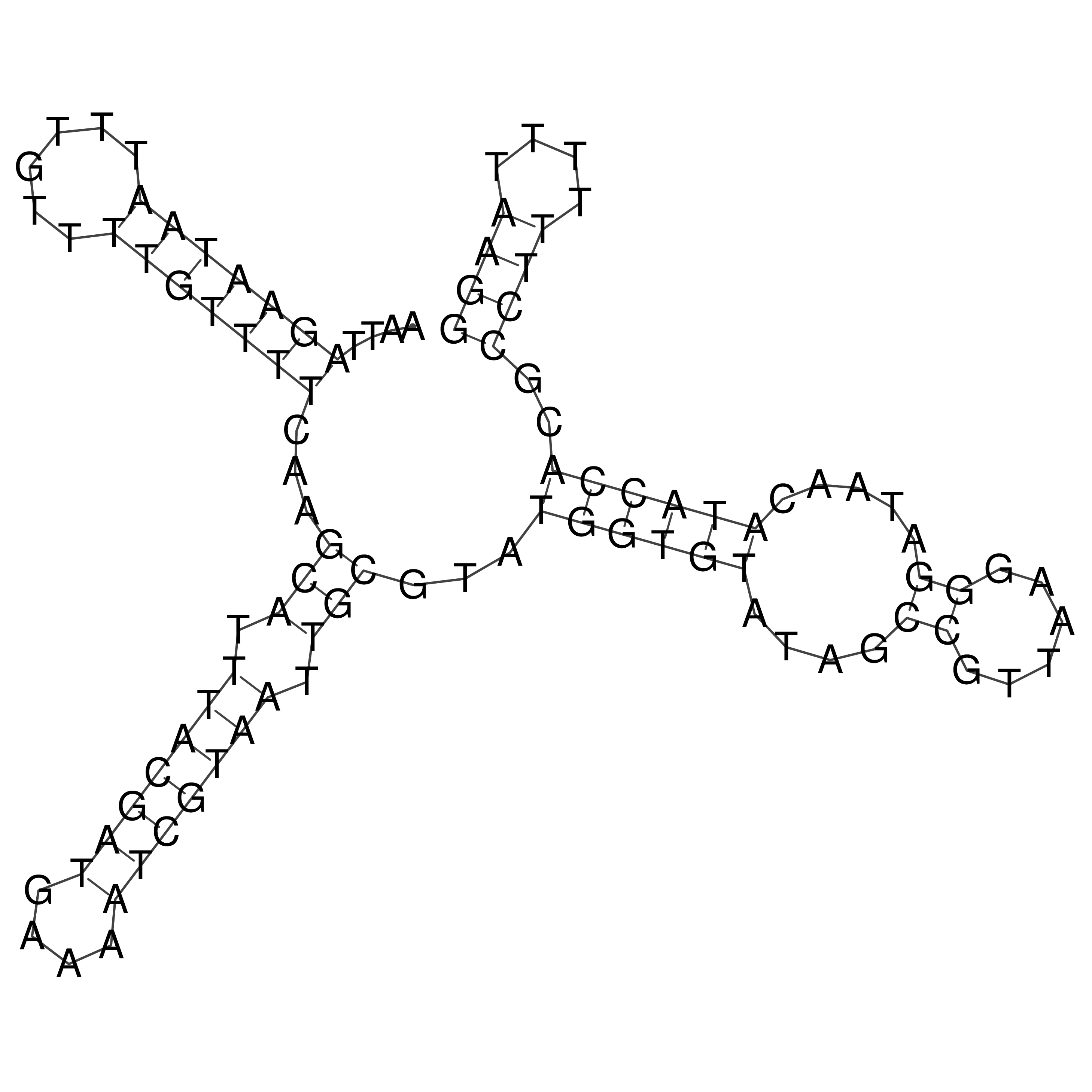
Relaxase
| ID | 4181 | GenBank | WP_277696281 |
| Name | mobF_LVQ68_RS27355_pKP037-3 |
UniProt ID | _ |
| Length | 969 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 969 a.a. Molecular weight: 108406.46 Da Isoelectric Point: 9.7611
>WP_277696281.1 MobF family relaxase [Klebsiella pneumoniae]
MLNITTITRQRVDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISVM
RKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVENT
NKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGYN
LRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYEG
WKNRAKELQIDFDSREWKGAANDDIARNTQPAALEKPIEYQADKVMTFAIRSLTERQSVITERELMDVAM
KHGYGRLTLDDIRAARERAITSGNLIKEEATYAAATTNKKQQNVAMTREQWVTELVKAGRNKDEARKLVD
KGITNGRLVQQKPRFTTQLAQQRERNILKMEREGRGKIQTPYTREFSEGWLASRTLKPEQLKAVMGIIHT
PNQFISVHGFAGTGKSYMTKSAADFLKEQGVHVTSLAPYGSQVKALQAEGLESRTLQSFLRASDKKIGPG
SVVFIDEAGVIPARQMEETMRVIRDAGARAVFLGDTKQTKAVEAGKPFEQLIKAGMETAYMKDIQRQKDP
ELLKAALNAAEGKIKASLTHVTSIVEEKNHDQRYHRIVGDYVAMPPTDRANALIITGTNDSRKKINAYIQ
AELGLKGQGINYPLLNRLDTTQAQRQHSKYYEKGAVIIPERDYSNGLKRGAVYTVLDTGPGNKLTVSDGS
GDTIAFSPARFSKLSVYSVEKTELAVGDQVRITRNDAHLDLANGDRFKVKGVQSGEVLLENEKGRLVKID
ANKPMYLGLAYASTVHSAQGLTCDKVFINMDTRSLTTAKDVYYVAVTRAKHEAVIYTDDEKKLDKAASRE
GFKTAALELEQLKRYAKEQQHDAREKGRDKTEPTQEKDNAKRKGKGHDKNNDERAFTSL
MLNITTITRQRVDKVVDYYADGADDYYAKDGNAMQWQGAGAEELGLTGEVEQKRFAKLLDGKVTDDISVM
RKTANSESKERLGYDLTFSAPKGVTLQALVNGDKRIIEAHDRAVTKAIEEAERLALGRFTVNKKTHVENT
NKLIVGKFRHETSRELDPDLHTHAFVMNLTKTSDGKWRSLTNDGIVNSLTLLGNVYKSELARELELAGYN
LRYDRNGTFDLAHFSQEQIAQFSQRSQQIEAELAKNGLSRETASHEQKNAAALKTRKSKGEVDRDVLYEG
WKNRAKELQIDFDSREWKGAANDDIARNTQPAALEKPIEYQADKVMTFAIRSLTERQSVITERELMDVAM
KHGYGRLTLDDIRAARERAITSGNLIKEEATYAAATTNKKQQNVAMTREQWVTELVKAGRNKDEARKLVD
KGITNGRLVQQKPRFTTQLAQQRERNILKMEREGRGKIQTPYTREFSEGWLASRTLKPEQLKAVMGIIHT
PNQFISVHGFAGTGKSYMTKSAADFLKEQGVHVTSLAPYGSQVKALQAEGLESRTLQSFLRASDKKIGPG
SVVFIDEAGVIPARQMEETMRVIRDAGARAVFLGDTKQTKAVEAGKPFEQLIKAGMETAYMKDIQRQKDP
ELLKAALNAAEGKIKASLTHVTSIVEEKNHDQRYHRIVGDYVAMPPTDRANALIITGTNDSRKKINAYIQ
AELGLKGQGINYPLLNRLDTTQAQRQHSKYYEKGAVIIPERDYSNGLKRGAVYTVLDTGPGNKLTVSDGS
GDTIAFSPARFSKLSVYSVEKTELAVGDQVRITRNDAHLDLANGDRFKVKGVQSGEVLLENEKGRLVKID
ANKPMYLGLAYASTVHSAQGLTCDKVFINMDTRSLTTAKDVYYVAVTRAKHEAVIYTDDEKKLDKAASRE
GFKTAALELEQLKRYAKEQQHDAREKGRDKTEPTQEKDNAKRKGKGHDKNNDERAFTSL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4200 | GenBank | WP_001001035 |
| Name | t4cp2_LVQ68_RS27360_pKP037-3 |
UniProt ID | _ |
| Length | 525 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 525 a.a. Molecular weight: 59019.93 Da Isoelectric Point: 9.5507
>WP_001001035.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacterales]
MNEADKGKICLGALLFTPLVTWFIAAKTLYGFTPKDARERLVLLIQQTFDLWPLWGALLVGEIIAIIGLI
IFFKFNHQIFKGAPFKKIYRGTRLVSQRALASLTKEREPQITIADIPIPTKAEGTHISIAGATGVGKSTI
FAEMMKGCLMRGKMQRVASKKDRMIILDPDGDFLSRFYKKGDKILNPYDARTEGWVFFNEIKSDFDFERF
GVSIVPTAKDSQTEEWNGYGRLLFTEVSRKVFNTSRNPTMEEVFYWTNEVPLEELEEYVRGTKAQALFAG
SGRAVGSARFVLSNKLAAHLKMPAGNFSIREWLEDPKGGNLYITWTDEMREALKSLISCWTDSIFSIVLG
MSKSNTRKIWTYIDELESLDYLPSLRDALTKGRKKGLRVVTGYQSYSQVISIYGSDVAETLLSNHRTSVV
MAAGRLGERTLEFVSKSLGEIEGEREKTGISRRFGQMGTKNINDDHKRERAVTPTEIATLDDLTGYIAFP
GNLPVAKFETHHVNYTRSNPVPGVVLRESTAFAGM
MNEADKGKICLGALLFTPLVTWFIAAKTLYGFTPKDARERLVLLIQQTFDLWPLWGALLVGEIIAIIGLI
IFFKFNHQIFKGAPFKKIYRGTRLVSQRALASLTKEREPQITIADIPIPTKAEGTHISIAGATGVGKSTI
FAEMMKGCLMRGKMQRVASKKDRMIILDPDGDFLSRFYKKGDKILNPYDARTEGWVFFNEIKSDFDFERF
GVSIVPTAKDSQTEEWNGYGRLLFTEVSRKVFNTSRNPTMEEVFYWTNEVPLEELEEYVRGTKAQALFAG
SGRAVGSARFVLSNKLAAHLKMPAGNFSIREWLEDPKGGNLYITWTDEMREALKSLISCWTDSIFSIVLG
MSKSNTRKIWTYIDELESLDYLPSLRDALTKGRKKGLRVVTGYQSYSQVISIYGSDVAETLLSNHRTSVV
MAAGRLGERTLEFVSKSLGEIEGEREKTGISRRFGQMGTKNINDDHKRERAVTPTEIATLDDLTGYIAFP
GNLPVAKFETHHVNYTRSNPVPGVVLRESTAFAGM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 24225..33854
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LVQ68_RS27465 (LVQ68_27425) | 19867..20751 | - | 885 | WP_023187082 | replication protein RepA | - |
| LVQ68_RS27470 (LVQ68_27430) | 20784..21368 | - | 585 | WP_021536796 | recombinase family protein | - |
| LVQ68_RS27475 (LVQ68_27435) | 21820..22065 | + | 246 | WP_021536799 | AbrB/MazE/SpoVT family DNA-binding domain-containing protein | - |
| LVQ68_RS27480 (LVQ68_27440) | 22077..22334 | + | 258 | WP_108714640 | hypothetical protein | - |
| LVQ68_RS27485 (LVQ68_27445) | 22324..22650 | + | 327 | WP_016240388 | endoribonuclease MazF | - |
| LVQ68_RS27490 (LVQ68_27450) | 22659..23096 | + | 438 | WP_060628251 | hypothetical protein | - |
| LVQ68_RS27495 (LVQ68_27455) | 23122..23358 | - | 237 | WP_042041953 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| LVQ68_RS27500 (LVQ68_27460) | 23524..24249 | - | 726 | WP_225386900 | restriction endonuclease | - |
| LVQ68_RS27505 (LVQ68_27465) | 24225..24539 | - | 315 | WP_023223516 | hypothetical protein | tfc15 |
| LVQ68_RS27510 (LVQ68_27470) | 24542..24859 | - | 318 | WP_108714639 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| LVQ68_RS27515 (LVQ68_27475) | 24865..25302 | - | 438 | WP_108714638 | hypothetical protein | - |
| LVQ68_RS27520 (LVQ68_27480) | 25314..25550 | - | 237 | WP_000089855 | H-NS histone family protein | - |
| LVQ68_RS27525 (LVQ68_27485) | 25613..26353 | + | 741 | WP_000815895 | lytic transglycosylase domain-containing protein | virB1 |
| LVQ68_RS27530 (LVQ68_27490) | 26376..26693 | + | 318 | WP_001267114 | hypothetical protein | - |
| LVQ68_RS27535 (LVQ68_27495) | 26677..26967 | + | 291 | WP_114266869 | TrbC/VirB2 family protein | virB2 |
| LVQ68_RS27540 (LVQ68_27500) | 27017..27331 | + | 315 | WP_001053888 | VirB3 family type IV secretion system protein | virB3 |
| LVQ68_RS27545 (LVQ68_27505) | 27331..29961 | + | 2631 | WP_108714637 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| LVQ68_RS27550 (LVQ68_27510) | 29971..30678 | + | 708 | WP_108714636 | type IV secretion system protein | virB5 |
| LVQ68_RS27555 (LVQ68_27515) | 30682..30918 | + | 237 | WP_000733633 | EexN family lipoprotein | - |
| LVQ68_RS27560 (LVQ68_27520) | 30937..31980 | + | 1044 | WP_021536808 | type IV secretion system protein | virB6 |
| LVQ68_RS27565 (LVQ68_27525) | 32075..32245 | + | 171 | WP_021536809 | hypothetical protein | - |
| LVQ68_RS27570 (LVQ68_27530) | 32248..32964 | + | 717 | WP_000914645 | type IV secretion system protein | virB8 |
| LVQ68_RS27575 (LVQ68_27535) | 32967..33854 | + | 888 | WP_000744355 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
Host bacterium
| ID | 6677 | GenBank | NZ_CP089873 |
| Plasmid name | pKP037-3 | Incompatibility group | - |
| Plasmid size | 34997 bp | Coordinate of oriT [Strand] | 7459..7559 [+] |
| Host baterium | Klebsiella pneumoniae strain KP037 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |