Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106227 |
| Name | oriT_pKP46-mcr8 |
| Organism | Klebsiella pneumoniae strain 46 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP088125 (63004..63053 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKP46-mcr8
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file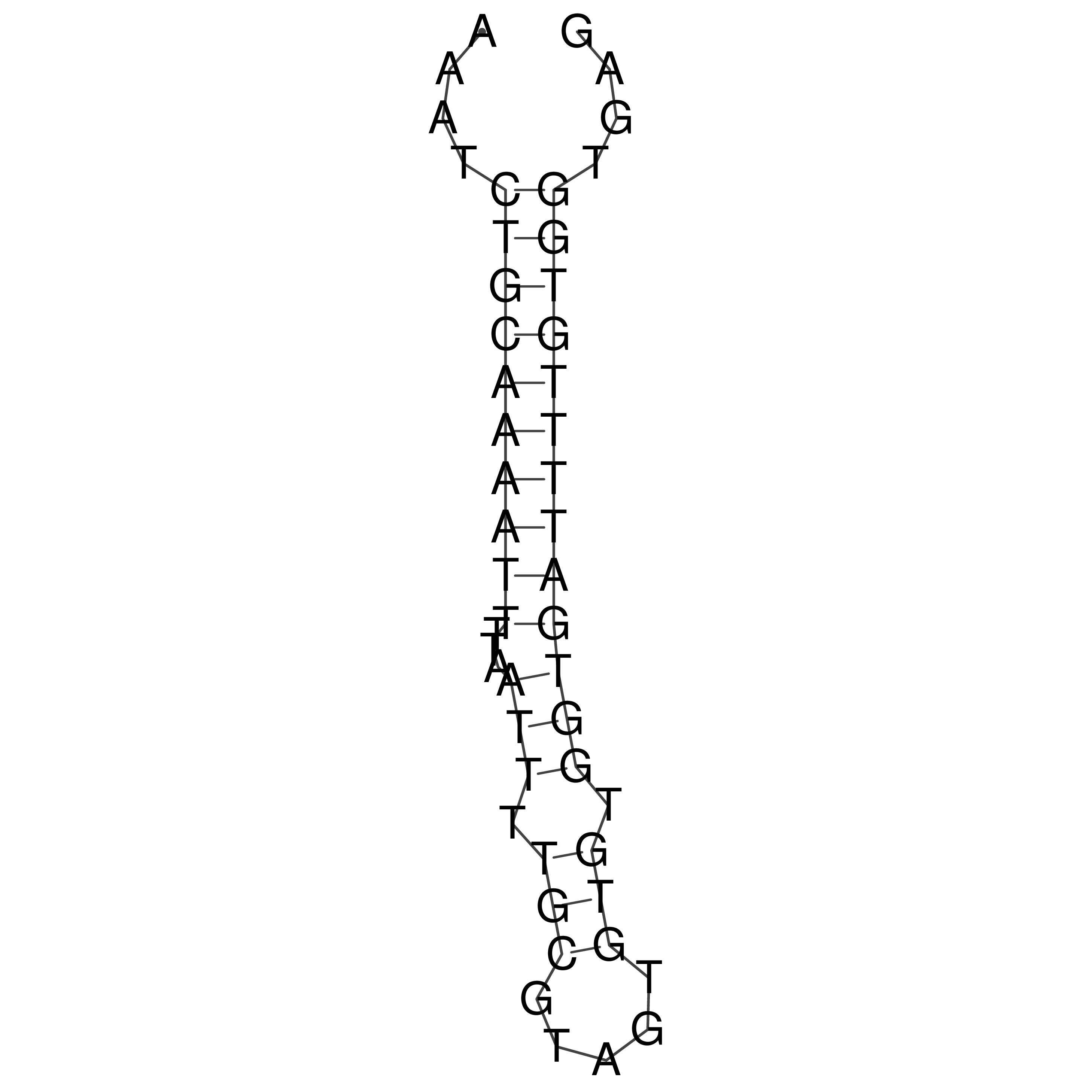
Relaxase
| ID | 4171 | GenBank | WP_172691966 |
| Name | traI_LRM33_RS29500_pKP46-mcr8 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190320.99 Da Isoelectric Point: 6.1403
>WP_172691966.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDHPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWEPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDTAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWMKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDHPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWEPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDTAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWMKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4188 | GenBank | WP_228303505 |
| Name | traD_LRM33_RS29495_pKP46-mcr8 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85885.99 Da Isoelectric Point: 5.0596
>WP_228303505.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVIAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVIAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 62446..90561
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| LRM33_RS29305 (LRM33_29305) | 58496..58876 | + | 381 | WP_020802391 | hypothetical protein | - |
| LRM33_RS29310 (LRM33_29310) | 58942..59289 | + | 348 | WP_032445771 | hypothetical protein | - |
| LRM33_RS29315 (LRM33_29315) | 59384..59530 | + | 147 | WP_004152750 | hypothetical protein | - |
| LRM33_RS29320 (LRM33_29320) | 59581..60414 | + | 834 | WP_032445769 | N-6 DNA methylase | - |
| LRM33_RS29325 (LRM33_29325) | 61230..62051 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| LRM33_RS29330 (LRM33_29330) | 62084..62413 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| LRM33_RS29335 (LRM33_29335) | 62446..62931 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| LRM33_RS29340 (LRM33_29340) | 63322..63738 | + | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| LRM33_RS29345 (LRM33_29345) | 63938..64657 | + | 720 | WP_172691949 | conjugal transfer protein TrbJ | - |
| LRM33_RS29350 (LRM33_29350) | 64790..64993 | + | 204 | WP_171486139 | TraY domain-containing protein | - |
| LRM33_RS29355 (LRM33_29355) | 65058..65426 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| LRM33_RS29360 (LRM33_29360) | 65440..65745 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| LRM33_RS29365 (LRM33_29365) | 65765..66331 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| LRM33_RS29370 (LRM33_29370) | 66318..67058 | + | 741 | WP_172691947 | type-F conjugative transfer system secretin TraK | traK |
| LRM33_RS29375 (LRM33_29375) | 67058..68482 | + | 1425 | WP_185775340 | F-type conjugal transfer pilus assembly protein TraB | traB |
| LRM33_RS29380 (LRM33_29380) | 68555..69139 | + | 585 | WP_040120377 | type IV conjugative transfer system lipoprotein TraV | traV |
| LRM33_RS29385 (LRM33_29385) | 69462..69680 | + | 219 | WP_004171484 | hypothetical protein | - |
| LRM33_RS29390 (LRM33_29390) | 69681..69998 | + | 318 | WP_114473059 | hypothetical protein | - |
| LRM33_RS29395 (LRM33_29395) | 70065..70469 | + | 405 | WP_100748549 | hypothetical protein | - |
| LRM33_RS29400 (LRM33_29400) | 70494..70892 | + | 399 | WP_167875005 | hypothetical protein | - |
| LRM33_RS29405 (LRM33_29405) | 70964..73603 | + | 2640 | WP_150388098 | type IV secretion system protein TraC | virb4 |
| LRM33_RS29410 (LRM33_29410) | 73603..73992 | + | 390 | WP_013214025 | type-F conjugative transfer system protein TrbI | - |
| LRM33_RS29415 (LRM33_29415) | 73992..74627 | + | 636 | WP_020325113 | type-F conjugative transfer system protein TraW | traW |
| LRM33_RS29420 (LRM33_29420) | 74662..75063 | + | 402 | WP_004194979 | hypothetical protein | - |
| LRM33_RS29425 (LRM33_29425) | 75060..76049 | + | 990 | WP_032441886 | conjugal transfer pilus assembly protein TraU | traU |
| LRM33_RS29430 (LRM33_29430) | 76062..76700 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| LRM33_RS29435 (LRM33_29435) | 76759..78714 | + | 1956 | WP_172691973 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| LRM33_RS29440 (LRM33_29440) | 78746..78982 | + | 237 | WP_114499720 | conjugal transfer protein TrbE | - |
| LRM33_RS29660 | 78979..79164 | + | 186 | WP_042922350 | hypothetical protein | - |
| LRM33_RS29445 (LRM33_29445) | 79210..79536 | + | 327 | WP_016529362 | hypothetical protein | - |
| LRM33_RS29450 (LRM33_29450) | 79557..80309 | + | 753 | WP_172691972 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| LRM33_RS29455 (LRM33_29455) | 80320..80559 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| LRM33_RS29460 (LRM33_29460) | 80531..81088 | + | 558 | WP_172691971 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| LRM33_RS29465 (LRM33_29465) | 81135..81581 | + | 447 | WP_172691970 | conjugal transfer protein TrbF | - |
| LRM33_RS29470 (LRM33_29470) | 81571..82938 | + | 1368 | WP_185775342 | conjugal transfer pilus assembly protein TraH | traH |
| LRM33_RS29475 (LRM33_29475) | 82938..85787 | + | 2850 | WP_172691976 | conjugal transfer mating-pair stabilization protein TraG | traG |
| LRM33_RS29480 (LRM33_29480) | 85793..86320 | + | 528 | WP_040120388 | conjugal transfer protein TraS | - |
| LRM33_RS29485 (LRM33_29485) | 86509..87240 | + | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| LRM33_RS29490 (LRM33_29490) | 87433..88122 | + | 690 | WP_172691968 | hypothetical protein | - |
| LRM33_RS29495 (LRM33_29495) | 88249..90561 | + | 2313 | WP_228303505 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 6664 | GenBank | NZ_CP088125 |
| Plasmid name | pKP46-mcr8 | Incompatibility group | IncFIA |
| Plasmid size | 101184 bp | Coordinate of oriT [Strand] | 63004..63053 [-] |
| Host baterium | Klebsiella pneumoniae strain 46 |
Cargo genes
| Drug resistance gene | mcr-8 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |