Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106211 |
| Name | oriT_pSW25tet_A |
| Organism | Klebsiella pneumoniae strain AHSWKP25 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP091051 (80918..80966 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pSW25tet_A
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file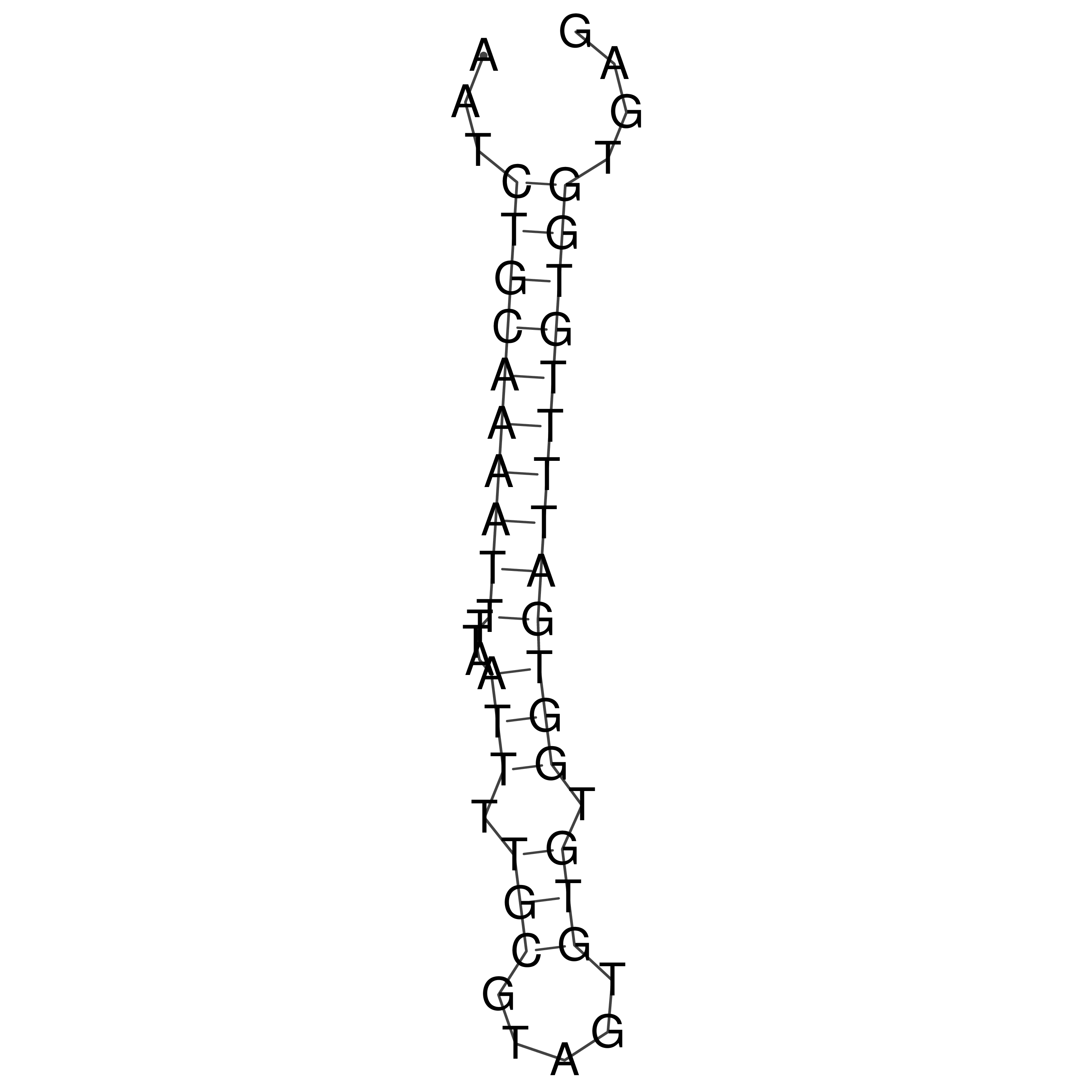
Relaxase
| ID | 4162 | GenBank | WP_126291924 |
| Name | traI_L1P15_RS28200_pSW25tet_A |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190438.05 Da Isoelectric Point: 5.9442
>WP_126291924.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTWIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
MQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQIPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGNSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRTLNGMIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRHQEAVIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTWIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
MQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQIPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGNSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRTLNGMIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIQHSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRHQEAVIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1856 | GenBank | WP_020805752 |
| Name | WP_020805752_pSW25tet_A |
UniProt ID | A0A377TIM3 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14772.81 Da Isoelectric Point: 4.5715
>WP_020805752.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Gammaproteobacteria]
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A377TIM3 |
T4CP
| ID | 4181 | GenBank | WP_065808185 |
| Name | traD_L1P15_RS28195_pSW25tet_A |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85734.63 Da Isoelectric Point: 4.9615
>WP_065808185.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIILYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTESHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIILYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTVWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTESHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKSPAAEPSVRATEPPVLRVTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 83345..110330
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| L1P15_RS28005 (L1P15_28005) | 79798..80328 | + | 531 | WP_023292160 | antirestriction protein | - |
| L1P15_RS28010 (L1P15_28010) | 80366..80773 | - | 408 | WP_223170089 | transglycosylase SLT domain-containing protein | - |
| L1P15_RS28015 (L1P15_28015) | 81277..81669 | + | 393 | WP_020805752 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| L1P15_RS28020 (L1P15_28020) | 81907..82608 | + | 702 | WP_223367236 | hypothetical protein | - |
| L1P15_RS28025 (L1P15_28025) | 82694..82894 | + | 201 | WP_048289787 | TraY domain-containing protein | - |
| L1P15_RS28030 (L1P15_28030) | 82963..83331 | + | 369 | WP_020316649 | type IV conjugative transfer system pilin TraA | - |
| L1P15_RS28035 (L1P15_28035) | 83345..83650 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| L1P15_RS28040 (L1P15_28040) | 83670..84236 | + | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| L1P15_RS28045 (L1P15_28045) | 84223..84963 | + | 741 | WP_013214019 | type-F conjugative transfer system secretin TraK | traK |
| L1P15_RS28050 (L1P15_28050) | 84963..86387 | + | 1425 | WP_126291926 | F-type conjugal transfer pilus assembly protein TraB | traB |
| L1P15_RS28055 (L1P15_28055) | 86380..86976 | + | 597 | WP_065808167 | conjugal transfer pilus-stabilizing protein TraP | - |
| L1P15_RS28060 (L1P15_28060) | 86999..87568 | + | 570 | WP_065808168 | type IV conjugative transfer system lipoprotein TraV | traV |
| L1P15_RS28065 (L1P15_28065) | 87700..88110 | + | 411 | WP_065808169 | hypothetical protein | - |
| L1P15_RS28070 (L1P15_28070) | 88115..88405 | + | 291 | WP_065808170 | hypothetical protein | - |
| L1P15_RS28075 (L1P15_28075) | 88429..88647 | + | 219 | WP_004171484 | hypothetical protein | - |
| L1P15_RS28080 (L1P15_28080) | 88648..88965 | + | 318 | WP_065808171 | hypothetical protein | - |
| L1P15_RS28085 (L1P15_28085) | 89032..89436 | + | 405 | WP_065808172 | hypothetical protein | - |
| L1P15_RS28090 (L1P15_28090) | 89479..89880 | + | 402 | WP_065808173 | hypothetical protein | - |
| L1P15_RS28095 (L1P15_28095) | 89888..90286 | + | 399 | WP_074423045 | hypothetical protein | - |
| L1P15_RS28100 (L1P15_28100) | 90357..92996 | + | 2640 | WP_065808175 | type IV secretion system protein TraC | virb4 |
| L1P15_RS28105 (L1P15_28105) | 92996..93385 | + | 390 | WP_020326931 | type-F conjugative transfer system protein TrbI | - |
| L1P15_RS28110 (L1P15_28110) | 93385..94011 | + | 627 | WP_065808176 | type-F conjugative transfer system protein TraW | traW |
| L1P15_RS28115 (L1P15_28115) | 94055..95014 | + | 960 | WP_234833367 | conjugal transfer pilus assembly protein TraU | traU |
| L1P15_RS28120 (L1P15_28120) | 95027..95665 | + | 639 | WP_065808178 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| L1P15_RS28125 (L1P15_28125) | 95662..96045 | + | 384 | WP_065808179 | hypothetical protein | - |
| L1P15_RS28130 (L1P15_28130) | 96042..97997 | + | 1956 | WP_065808195 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| L1P15_RS28135 (L1P15_28135) | 98030..98311 | + | 282 | WP_022644736 | hypothetical protein | - |
| L1P15_RS28140 (L1P15_28140) | 98301..98528 | + | 228 | WP_065808180 | conjugal transfer protein TrbE | - |
| L1P15_RS28145 (L1P15_28145) | 98539..98865 | + | 327 | WP_065808181 | hypothetical protein | - |
| L1P15_RS28150 (L1P15_28150) | 98886..99638 | + | 753 | WP_065808182 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| L1P15_RS28155 (L1P15_28155) | 100094..100333 | + | 240 | WP_023316407 | type-F conjugative transfer system pilin chaperone TraQ | - |
| L1P15_RS28160 (L1P15_28160) | 100305..100862 | + | 558 | WP_020804677 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| L1P15_RS28165 (L1P15_28165) | 100908..101351 | + | 444 | WP_023340929 | F-type conjugal transfer protein TrbF | - |
| L1P15_RS28170 (L1P15_28170) | 101329..102708 | + | 1380 | WP_072198238 | conjugal transfer pilus assembly protein TraH | traH |
| L1P15_RS28175 (L1P15_28175) | 102708..105557 | + | 2850 | WP_065808184 | conjugal transfer mating-pair stabilization protein TraG | traG |
| L1P15_RS28180 (L1P15_28180) | 105560..106090 | + | 531 | WP_074423050 | conjugal transfer protein TraS | - |
| L1P15_RS28185 (L1P15_28185) | 106280..107011 | + | 732 | WP_032329926 | conjugal transfer complement resistance protein TraT | - |
| L1P15_RS28190 (L1P15_28190) | 107203..107892 | + | 690 | WP_074423046 | hypothetical protein | - |
| L1P15_RS28195 (L1P15_28195) | 108021..110330 | + | 2310 | WP_065808185 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 6648 | GenBank | NZ_CP091051 |
| Plasmid name | pSW25tet_A | Incompatibility group | IncFIA |
| Plasmid size | 120990 bp | Coordinate of oriT [Strand] | 80918..80966 [-] |
| Host baterium | Klebsiella pneumoniae strain AHSWKP25 |
Cargo genes
| Drug resistance gene | qnrS1, blaLAP-2, tet(A), floR, sul2, dfrA14 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |