Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106206 |
| Name | oriT_pW49-2-a |
| Organism | Escherichia marmotae strain W49-2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP093240 (49231..49314 [+], 84 nt) |
| oriT length | 84 nt |
| IRs (inverted repeats) | 28..33, 37..42 (GCCATC..GATGGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 84 nt
>oriT_pW49-2-a
ACGGGACAGGATATGTTTTGTTGAACCGCCATCTTCGATGGCCGCTCAACAAAACCCCTGGTCAGGGCGCAGCCCCGACACCCC
ACGGGACAGGATATGTTTTGTTGAACCGCCATCTTCGATGGCCGCTCAACAAAACCCCTGGTCAGGGCGCAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file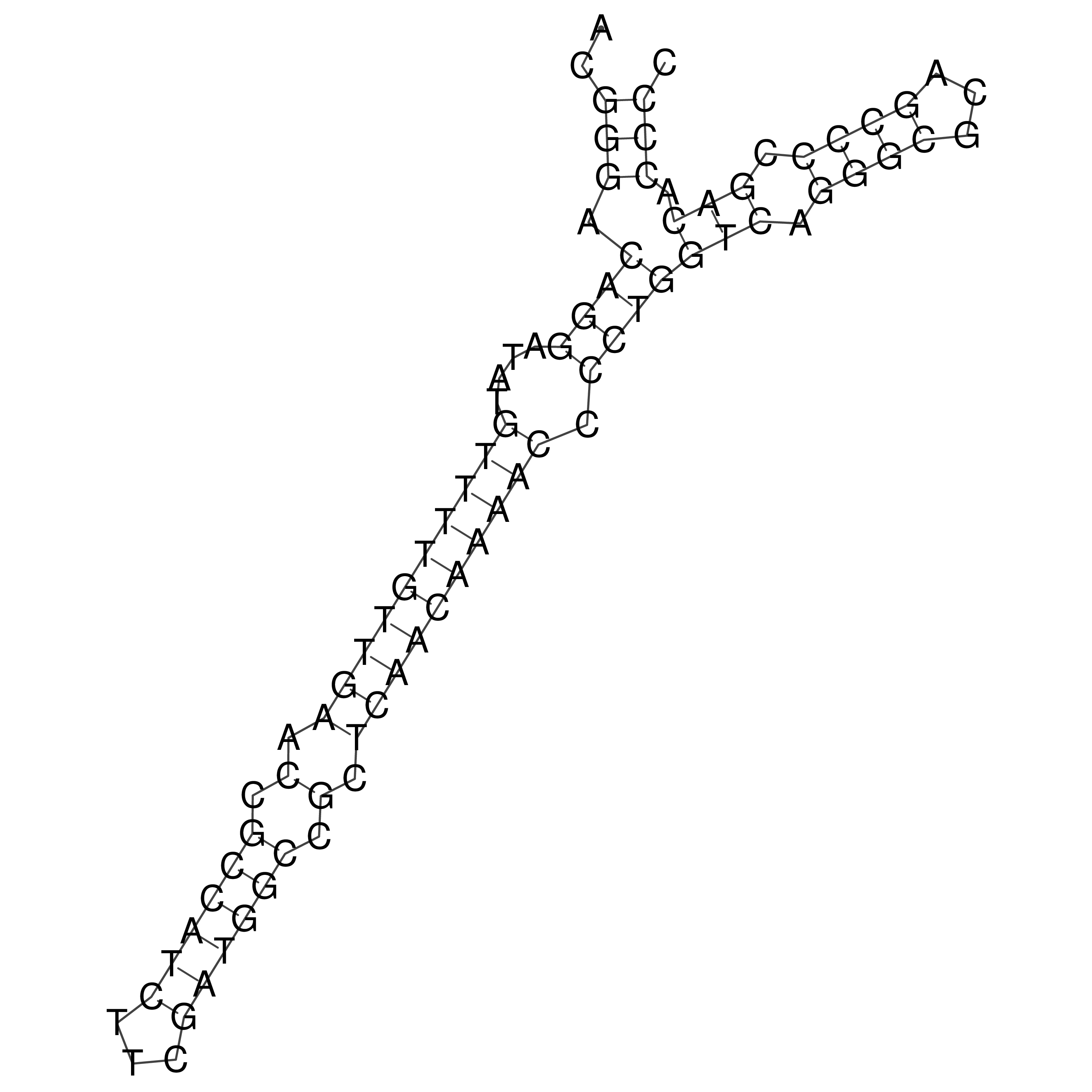
Relaxase
| ID | 4157 | GenBank | WP_241556898 |
| Name | Relaxase_MNY74_RS24790_pW49-2-a |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 644 a.a. Molecular weight: 74968.88 Da Isoelectric Point: 6.4714
>WP_241556898.1 TraI/MobA(P) family conjugative relaxase [Escherichia marmotae]
MDKKPGELLSPDNPYVRPVRSREAIVEHLGQFIEEGMDLYRNHVLSRSDDGRQQVLCGEVLCETNCFGIE
TAAAEMDAVASQNRRCQDPVYHAILSWREEDNPTNEQIFECARYALKALNMSDHQYVFAIHRDTEHVHCH
ITVNRINPKTYRAASLYDDYFRLHKACRELEQKYGFTPDNGCCQKDENGNWIRKQEEFKSIPRKARQMEY
HADKESLYSYAVDEARHTIGKILHSGEATWEALHTELIRLGLELKEKGEGLAVYSRHDDTVTPIKASTLH
PDLTKRVLEPYLGKFTPSPVVENYFDENDNYLGSNYPQEYVYDVRAHKRDIGASSARREERADAREALKA
RYNAYRAGWTRPEMDRDEIRARFYHVSGQFAWRRAQVRAEIRDPLLRKLAYNVLALERRQAMIALKETLR
EERKAFYARPENRRMGYWQWVEQQALNNDAAAISCLRGRFYMQKKMQQENRLSENAIVCAVADDIRPYEI
EGYQTQITRDGRIQYCQDGEVKLQDTGDRIEIADPYSQDGQHIAGAMVLAEVKSGERMYFAGEPQFVNKA
CSIVPWFNEGSEKPLPLTDRAQRQMAGYDTAPASPQKDEPEIAWESLRWLMEGAPKNPVQEDVRQQQKHD
YVHDDNYSSRYRRH
MDKKPGELLSPDNPYVRPVRSREAIVEHLGQFIEEGMDLYRNHVLSRSDDGRQQVLCGEVLCETNCFGIE
TAAAEMDAVASQNRRCQDPVYHAILSWREEDNPTNEQIFECARYALKALNMSDHQYVFAIHRDTEHVHCH
ITVNRINPKTYRAASLYDDYFRLHKACRELEQKYGFTPDNGCCQKDENGNWIRKQEEFKSIPRKARQMEY
HADKESLYSYAVDEARHTIGKILHSGEATWEALHTELIRLGLELKEKGEGLAVYSRHDDTVTPIKASTLH
PDLTKRVLEPYLGKFTPSPVVENYFDENDNYLGSNYPQEYVYDVRAHKRDIGASSARREERADAREALKA
RYNAYRAGWTRPEMDRDEIRARFYHVSGQFAWRRAQVRAEIRDPLLRKLAYNVLALERRQAMIALKETLR
EERKAFYARPENRRMGYWQWVEQQALNNDAAAISCLRGRFYMQKKMQQENRLSENAIVCAVADDIRPYEI
EGYQTQITRDGRIQYCQDGEVKLQDTGDRIEIADPYSQDGQHIAGAMVLAEVKSGERMYFAGEPQFVNKA
CSIVPWFNEGSEKPLPLTDRAQRQMAGYDTAPASPQKDEPEIAWESLRWLMEGAPKNPVQEDVRQQQKHD
YVHDDNYSSRYRRH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4177 | GenBank | WP_193803150 |
| Name | trbC_MNY74_RS24985_pW49-2-a |
UniProt ID | _ |
| Length | 738 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 738 a.a. Molecular weight: 83684.13 Da Isoelectric Point: 7.2887
>WP_193803150.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Escherichia]
MAIDSRPVDKQRVQHVIKSTWLMDILQSPFSLQAGMLGGLLVCLIAPWLLLPCLICAVIWLLVFTDQRFR
LPLRMPTDIGGFDYSTEREEANEFTLPGGFFRFIRRKLTVEKAGGIMCLGYARKRFLGRELWLTRDDCLR
HMQLLATTGSGKTEALLSLQLNAICLGRGYCYSDGKAETALAFSSWSLLRRFGLEDNFYVLNFLNGGRDR
FDELLSNDRTRPQSNTINLSNNATASYIIQLMDSMLPKVDGSSADWQDKARPMLYGLVYSLHYKCRRENL
RFSLAMLHKHLPLKEMAALYIEGKQNGWHQEGINALENYLSTLAGFQMDLVSRPAEWDQGVFDQHGFLIQ
QFSKMLAMFNDVYGHIFANDAGDIDLRDVLHNDRALVVLIPALELSKSESASLGKLYISAKRMVIARDLG
YHLEGKREEVLTAKKYRNRFPFPDIHDELGSYFAPGMDDLAAQMRSLGYMLIISAQDIQRFIAQFRGEYQ
TVNANLLVKWFMALQDEKDTYELARSTAGKDYFAELGEVKRSPGLFGNQFEDATTTHIREKDRISLDELK
NLNPGEGFISFKSALVPCNAVYIPDDEKLSSKLQMRINRFIDIGRPSESELVKKNPSLARLLPPDSQEVS
LVLQRTDEIMAGMAPRNPPPVVDPILARLAEITLNLDNRCDISYTPEQRGILLFEAAREAMKRSRGRWFL
AQTRHRPISVSDQVAHKLATDSTAFPSPVNTDKGTDPQ
MAIDSRPVDKQRVQHVIKSTWLMDILQSPFSLQAGMLGGLLVCLIAPWLLLPCLICAVIWLLVFTDQRFR
LPLRMPTDIGGFDYSTEREEANEFTLPGGFFRFIRRKLTVEKAGGIMCLGYARKRFLGRELWLTRDDCLR
HMQLLATTGSGKTEALLSLQLNAICLGRGYCYSDGKAETALAFSSWSLLRRFGLEDNFYVLNFLNGGRDR
FDELLSNDRTRPQSNTINLSNNATASYIIQLMDSMLPKVDGSSADWQDKARPMLYGLVYSLHYKCRRENL
RFSLAMLHKHLPLKEMAALYIEGKQNGWHQEGINALENYLSTLAGFQMDLVSRPAEWDQGVFDQHGFLIQ
QFSKMLAMFNDVYGHIFANDAGDIDLRDVLHNDRALVVLIPALELSKSESASLGKLYISAKRMVIARDLG
YHLEGKREEVLTAKKYRNRFPFPDIHDELGSYFAPGMDDLAAQMRSLGYMLIISAQDIQRFIAQFRGEYQ
TVNANLLVKWFMALQDEKDTYELARSTAGKDYFAELGEVKRSPGLFGNQFEDATTTHIREKDRISLDELK
NLNPGEGFISFKSALVPCNAVYIPDDEKLSSKLQMRINRFIDIGRPSESELVKKNPSLARLLPPDSQEVS
LVLQRTDEIMAGMAPRNPPPVVDPILARLAEITLNLDNRCDISYTPEQRGILLFEAAREAMKRSRGRWFL
AQTRHRPISVSDQVAHKLATDSTAFPSPVNTDKGTDPQ
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 86035..115544
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MNY74_RS24980 (MNY74_24980) | 83299..83799 | - | 501 | WP_241556901 | phospholipase D family protein | - |
| MNY74_RS24985 (MNY74_24985) | 83829..86045 | - | 2217 | WP_193803150 | F-type conjugative transfer protein TrbC | - |
| MNY74_RS24990 (MNY74_24990) | 86035..86955 | - | 921 | WP_193803149 | conjugal transfer protein TraF | trbB |
| MNY74_RS24995 (MNY74_24995) | 86965..88098 | - | 1134 | WP_241556879 | conjugal transfer protein TrbA | trbA |
| MNY74_RS25000 (MNY74_25000) | 88114..88896 | - | 783 | WP_001317493 | IS21-like element IS100kyp family helper ATPase IstB | - |
| MNY74_RS25005 (MNY74_25005) | 88893..89915 | - | 1023 | WP_000255956 | IS21-like element IS100 family transposase | - |
| MNY74_RS25010 (MNY74_25010) | 89929..90297 | - | 369 | WP_241556880 | hypothetical protein | trbA |
| MNY74_RS25015 (MNY74_25015) | 90584..90721 | - | 138 | WP_107135979 | Hok/Gef family protein | - |
| MNY74_RS25020 (MNY74_25020) | 91081..91734 | - | 654 | WP_105289555 | DUF5710 domain-containing protein | - |
| MNY74_RS25025 (MNY74_25025) | 91774..92130 | - | 357 | WP_107135981 | hypothetical protein | - |
| MNY74_RS25030 (MNY74_25030) | 92384..92584 | - | 201 | WP_097468676 | hemolysin expression modulator Hha | - |
| MNY74_RS25035 (MNY74_25035) | 92647..93183 | - | 537 | WP_170898668 | hypothetical protein | - |
| MNY74_RS25040 (MNY74_25040) | 93323..93871 | - | 549 | WP_241556881 | hypothetical protein | - |
| MNY74_RS25045 (MNY74_25045) | 93886..96021 | - | 2136 | WP_193803147 | DotA/TraY family protein | traY |
| MNY74_RS25050 (MNY74_25050) | 96105..96449 | - | 345 | WP_215251741 | hypothetical protein | - |
| MNY74_RS25055 (MNY74_25055) | 96451..96951 | - | 501 | WP_193803146 | hypothetical protein | - |
| MNY74_RS25060 (MNY74_25060) | 97118..97411 | - | 294 | WP_063848353 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| MNY74_RS25065 (MNY74_25065) | 97411..97683 | - | 273 | WP_063848352 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
| MNY74_RS25070 (MNY74_25070) | 97820..97966 | - | 147 | WP_181475174 | hypothetical protein | - |
| MNY74_RS25075 (MNY74_25075) | 98111..98641 | - | 531 | WP_170898664 | conjugal transfer protein TraX | - |
| MNY74_RS25080 (MNY74_25080) | 98638..99858 | - | 1221 | WP_001039317 | conjugal transfer protein TraW | traW |
| MNY74_RS25085 (MNY74_25085) | 99882..102938 | - | 3057 | WP_241556882 | ATP-binding protein | traU |
| MNY74_RS25090 (MNY74_25090) | 102942..103553 | - | 612 | WP_241556883 | hypothetical protein | traT |
| MNY74_RS25095 (MNY74_25095) | 103547..103816 | - | 270 | WP_241556884 | hypothetical protein | - |
| MNY74_RS25100 (MNY74_25100) | 103872..104279 | - | 408 | WP_001059622 | DUF6750 family protein | traR |
| MNY74_RS25105 (MNY74_25105) | 104311..104835 | - | 525 | WP_241556902 | conjugal transfer protein TraQ | traQ |
| MNY74_RS25110 (MNY74_25110) | 104846..105547 | - | 702 | WP_241556885 | conjugal transfer protein TraP | traP |
| MNY74_RS25115 (MNY74_25115) | 105548..106741 | - | 1194 | WP_241556886 | conjugal transfer protein TraO | traO |
| MNY74_RS25120 (MNY74_25120) | 106744..107769 | - | 1026 | WP_193803144 | DotH/IcmK family type IV secretion protein | traN |
| MNY74_RS25125 (MNY74_25125) | 107766..108437 | - | 672 | WP_050939885 | DotI/IcmL/TraM family protein | traM |
| MNY74_RS25130 (MNY74_25130) | 108457..108936 | - | 480 | WP_170898655 | hypothetical protein | traL |
| MNY74_RS25135 (MNY74_25135) | 108951..112394 | - | 3444 | WP_241556887 | DUF5710 domain-containing protein | - |
| MNY74_RS25140 (MNY74_25140) | 112412..112672 | - | 261 | WP_050939877 | IcmT/TraK family protein | traK |
| MNY74_RS25145 (MNY74_25145) | 112665..113837 | - | 1173 | WP_241556888 | plasmid transfer ATPase TraJ | virB11 |
| MNY74_RS25150 (MNY74_25150) | 113843..114625 | - | 783 | WP_105289544 | type IV secretory system conjugative DNA transfer family protein | traI |
| MNY74_RS25155 (MNY74_25155) | 114622..115047 | - | 426 | WP_157945705 | DotD/TraH family lipoprotein | - |
| MNY74_RS25160 (MNY74_25160) | 115074..115544 | - | 471 | WP_157917732 | lytic transglycosylase domain-containing protein | virB1 |
| MNY74_RS25165 (MNY74_25165) | 115544..116041 | - | 498 | WP_105289542 | hypothetical protein | - |
| MNY74_RS25170 (MNY74_25170) | 116051..116293 | - | 243 | WP_105289541 | hypothetical protein | - |
| MNY74_RS25175 (MNY74_25175) | 116432..116632 | - | 201 | WP_105289540 | hypothetical protein | - |
| MNY74_RS25180 (MNY74_25180) | 116622..117299 | - | 678 | WP_241556889 | hypothetical protein | - |
| MNY74_RS25185 (MNY74_25185) | 117397..117909 | - | 513 | WP_105289538 | transcription termination/antitermination NusG family protein | - |
Host bacterium
| ID | 6643 | GenBank | NZ_CP093240 |
| Plasmid name | pW49-2-a | Incompatibility group | IncI1 |
| Plasmid size | 118621 bp | Coordinate of oriT [Strand] | 49231..49314 [+] |
| Host baterium | Escherichia marmotae strain W49-2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | faeC, faeD, faeE, faeF, faeH, faeI, faeJ, papD, papC, papH |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |