Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106094 |
| Name | oriT_pPP1-1 |
| Organism | Pseudomonas syringae pv. pisi str. PP1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP034079 (56510..56686 [+], 177 nt) |
| oriT length | 177 nt |
| IRs (inverted repeats) | 76..83, 87..94 (CAAAGGGG..CCCCTTTG) 38..43, 52..57 (AAAAAG..CTTTTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 177 nt
>oriT_pPP1-1
CGAATTCGCAAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTATCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAAAGCGTTGTTGTTGTCGTTGTTGGTTGTCGTCATTCCAGGTGCCCCCAGGAGGTGATCTTGTGGC
CGAATTCGCAAAACAGGCACGTTACCGATTGTCTACAAAAAAGAGCTTCGCCTTTTTCGTATCCAATCGACGTGCCAAAGGGGATACCCCTTTGAAACCCCGCAGAGCGGGAAAGCGTTGTTGTTGTCGTTGTTGGTTGTCGTCATTCCAGGTGCCCCCAGGAGGTGATCTTGTGGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file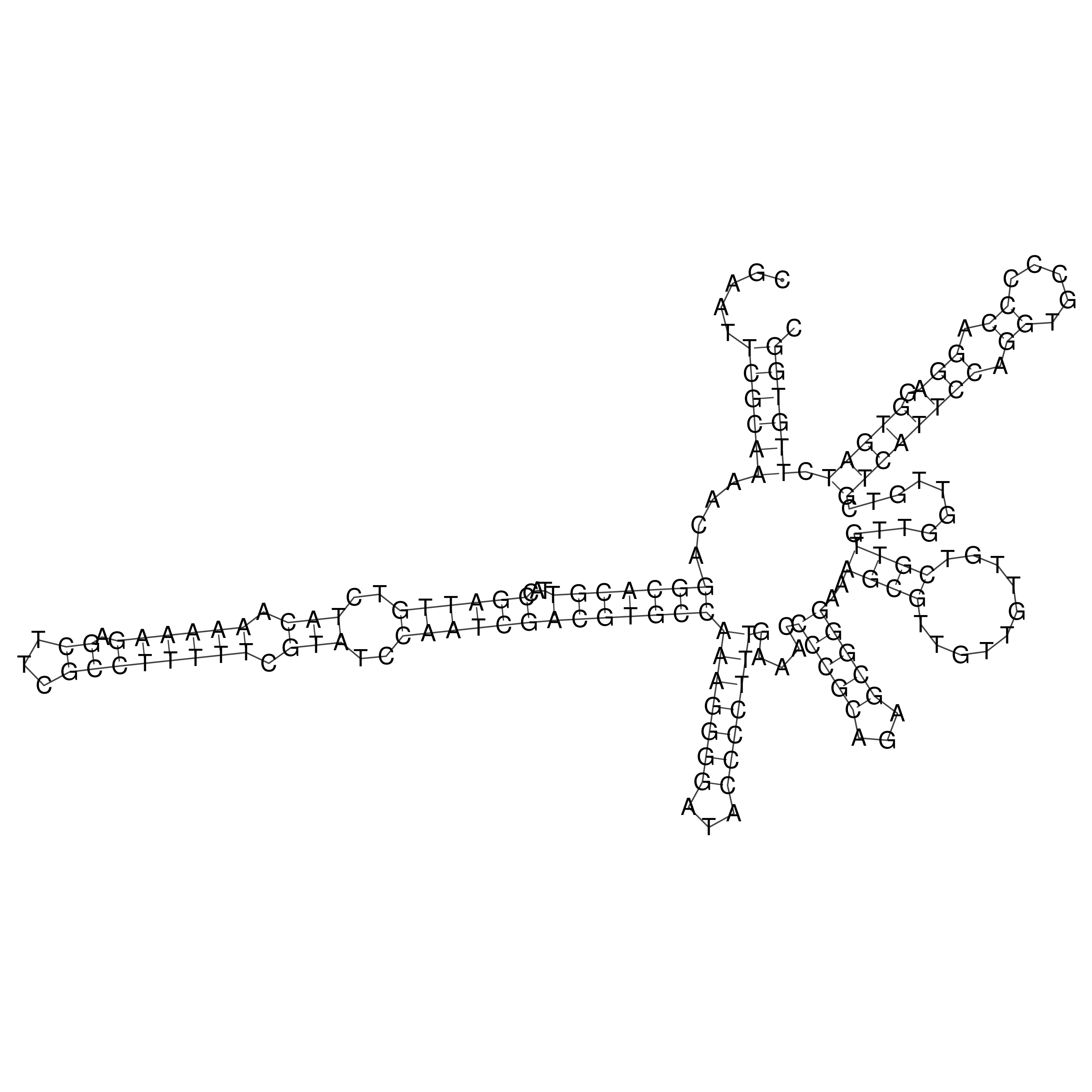
Relaxase
| ID | 4087 | GenBank | WP_124733764 |
| Name | Relaxase_N032_RS27570_pPP1-1 |
UniProt ID | _ |
| Length | 1262 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1262 a.a. Molecular weight: 139268.85 Da Isoelectric Point: 9.2151
>WP_124733764.1 LPD7 domain-containing protein [Pseudomonas syringae]
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDMKTGDVIERKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQDLVKQVIER
GVTTRADFYALVAEHGETRIRNEGKDTEYISVKLPGDAKGTNLKDTIFQDDFIVRRELKKPPLEASVIQE
RLLAWPQRAREIKYVNKATPKFRKQYSEASPEDRVRLLADREAKFYQVHGEHNDNVHTGQRQRDHQRSSA
ETAGRRTSAPADGLQDLSVSHVADHRQAGSARSRDGAVLLPSDAHIHVGQSQPGGDSGLRSSVPGGGRGR
GSRSTAERGRGGSGTAAVSQETTGTATPTGATGRRRAGAGKPRNARVRAGRVVPPYAKNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRAPTVASVPKAATATATPGRSTAGRRPRTSDKPRPPRQWRPGSVVPPYA
KNPHRVATVADIEQRARMLFDPLKRPTDKALVFKRASIKALTVNRQASTVAAYFTREAQHNQIAPAHRRA
IRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHNPKPNIEVNLMGSAAIRNLLDDEKE
DPGFSISGARGPGPEGVRDRVKRVMDRLAKQLDPAAASERARELSAKDLYTRKAKFSQNVHYLDKQTDKT
LFVDTGTAISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKAMNQSL
ADRRAELEIERGGLSIGPANDLTRHVNDSTRDVRDQADRMGVTVRIEALYGEGKTAEQVSQALTEQLDTV
PESERVAFVETVAITLGIPERGQPKGDQAFAQWQAQRAQPAADSTAPATAETQTSAPEPVSTPAATTPAT
APVSPEPVANSTAPATAKTPASVNDPDLQSPSELVRLEAQWRRDFPMSEADVKASDVVMGLRGEDHAVWI
ISTNDKTPEAAAMLTAYMENDSYREAFKASIVAAYKQVENSPKLVDDLDHLTEMAAQLVNEVEDRLFPAP
QAATGQTAPSRSNVIDGTLIEHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEKAMNAADLKQGDP
VRLEDLGTQPVVVQVIEEDGTVTNKTVNRREWSAQPVAPDREVAETTPKGQAEAAGTPELSSPDEEDGMG
MD
MLIRVSGYNTGAQEYLEQGNKSGREFTRDELDHRLIIEGQLSVTRAIYESIPDHGQDRYLTFTLSFKEDT
VSPELLKAVTTDFKNFFMHAYKPEEFNLYAEAHLPKMKTVTDMKTGDVIERKPHIHIIIPRINLLSGNEA
NPVDVYKNHEKYFEAFQEHINQKYGLSSPRENVRADITDAASVLSRYKGDDFYGKNRQFKQDLVKQVIER
GVTTRADFYALVAEHGETRIRNEGKDTEYISVKLPGDAKGTNLKDTIFQDDFIVRRELKKPPLEASVIQE
RLLAWPQRAREIKYVNKATPKFRKQYSEASPEDRVRLLADREAKFYQVHGEHNDNVHTGQRQRDHQRSSA
ETAGRRTSAPADGLQDLSVSHVADHRQAGSARSRDGAVLLPSDAHIHVGQSQPGGDSGLRSSVPGGGRGR
GSRSTAERGRGGSGTAAVSQETTGTATPTGATGRRRAGAGKPRNARVRAGRVVPPYAKNPHRVATIADIE
ERGRRLFDPLKRPSDNALVFYRAPTVASVPKAATATATPGRSTAGRRPRTSDKPRPPRQWRPGSVVPPYA
KNPHRVATVADIEQRARMLFDPLKRPTDKALVFKRASIKALTVNRQASTVAAYFTREAQHNQIAPAHRRA
IRRIDQQYFALRRAVFSDQRLTRQDKAQLVSVLTFERLKAREQFHNPKPNIEVNLMGSAAIRNLLDDEKE
DPGFSISGARGPGPEGVRDRVKRVMDRLAKQLDPAAASERARELSAKDLYTRKAKFSQNVHYLDKQTDKT
LFVDTGTAISMRRTGITEAGVSVALQLARERFGSTLTINGTAEFKKLVIEAVAKNGLDVHFTDKAMNQSL
ADRRAELEIERGGLSIGPANDLTRHVNDSTRDVRDQADRMGVTVRIEALYGEGKTAEQVSQALTEQLDTV
PESERVAFVETVAITLGIPERGQPKGDQAFAQWQAQRAQPAADSTAPATAETQTSAPEPVSTPAATTPAT
APVSPEPVANSTAPATAKTPASVNDPDLQSPSELVRLEAQWRRDFPMSEADVKASDVVMGLRGEDHAVWI
ISTNDKTPEAAAMLTAYMENDSYREAFKASIVAAYKQVENSPKLVDDLDHLTEMAAQLVNEVEDRLFPAP
QAATGQTAPSRSNVIDGTLIEHGEAPYQHNDDNQMSYFVTLKPEGGKPRTVWGVGLEKAMNAADLKQGDP
VRLEDLGTQPVVVQVIEEDGTVTNKTVNRREWSAQPVAPDREVAETTPKGQAEAAGTPELSSPDEEDGMG
MD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4102 | GenBank | WP_024650172 |
| Name | t4cp2_N032_RS27525_pPP1-1 |
UniProt ID | _ |
| Length | 557 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 557 a.a. Molecular weight: 61668.26 Da Isoelectric Point: 6.2657
>WP_024650172.1 type IV secretory system conjugative DNA transfer family protein [Pseudomonas syringae]
MAKAKSKPISPDNPQERIEEHPAILIGKHPTKDTFLASYGQTFVMLAAPPGTGKTVGVVTPNLLSYPDSV
VVNDPKFENWRDTAGFRAAAGHKVYRFSPELLETHRWNPLSALSRDPLYRLGQIRTLAGVLFVSDNPKNQ
EWYNKAANVFAAILLYLMEMEGMKLNGMKLTLPQAYEVASLGTGLGVWAQQAIEQHSTGPNALSVETLRE
LNGVFEASKNKSSGWSTTVDILRGALSMYAEKTVAWAVSDTDIDFTKLRKEKISIYFCVTGNAIKKYGPL
MNLFFTQAIQQNTDVMPEQGGHCADGTLILKYQVLFVIDEIAVMGRIEIMEMAPALTRGAGLRYIIIFQG
KDQLRSERTYGREAGNGIMQAFHIEIVYGTGNVELAKEYSTRLGNTTVRVHNQSLNRQKHEIGARGQTDS
YSEQPRPLLLPQEVSALPYGEELIFVEATKTTPAANIRARKIFWYEEPVFKERAGMPTPDVPSGDASKID
ALTVPVRTVEAKVAVADTKPMQAEQRQRWNPKEKAANAVPDQPDAAQPVDVEPDLEPAQTDDTPETM
MAKAKSKPISPDNPQERIEEHPAILIGKHPTKDTFLASYGQTFVMLAAPPGTGKTVGVVTPNLLSYPDSV
VVNDPKFENWRDTAGFRAAAGHKVYRFSPELLETHRWNPLSALSRDPLYRLGQIRTLAGVLFVSDNPKNQ
EWYNKAANVFAAILLYLMEMEGMKLNGMKLTLPQAYEVASLGTGLGVWAQQAIEQHSTGPNALSVETLRE
LNGVFEASKNKSSGWSTTVDILRGALSMYAEKTVAWAVSDTDIDFTKLRKEKISIYFCVTGNAIKKYGPL
MNLFFTQAIQQNTDVMPEQGGHCADGTLILKYQVLFVIDEIAVMGRIEIMEMAPALTRGAGLRYIIIFQG
KDQLRSERTYGREAGNGIMQAFHIEIVYGTGNVELAKEYSTRLGNTTVRVHNQSLNRQKHEIGARGQTDS
YSEQPRPLLLPQEVSALPYGEELIFVEATKTTPAANIRARKIFWYEEPVFKERAGMPTPDVPSGDASKID
ALTVPVRTVEAKVAVADTKPMQAEQRQRWNPKEKAANAVPDQPDAAQPVDVEPDLEPAQTDDTPETM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 33810..44164
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| N032_RS27405 (N032_27405) | 30199..30837 | - | 639 | WP_024647993 | recombinase family protein | - |
| N032_RS27410 (N032_27410) | 31128..31760 | + | 633 | WP_003381804 | ParA family protein | - |
| N032_RS27415 (N032_27415) | 31796..32050 | + | 255 | WP_003381801 | hypothetical protein | - |
| N032_RS27420 (N032_27420) | 32475..33140 | - | 666 | WP_024647998 | LexA family transcriptional regulator | - |
| N032_RS27425 (N032_27425) | 33228..33458 | + | 231 | WP_010215521 | Cro/CI family transcriptional regulator | - |
| N032_RS27430 (N032_27430) | 33810..34508 | + | 699 | WP_024650160 | lytic transglycosylase domain-containing protein | virB1 |
| N032_RS27435 (N032_27435) | 34508..34843 | + | 336 | WP_024650161 | hypothetical protein | - |
| N032_RS27440 (N032_27440) | 34856..35131 | + | 276 | WP_024650162 | VirB3 family type IV secretion system protein | virB3 |
| N032_RS27445 (N032_27445) | 35192..37744 | + | 2553 | WP_024650163 | VirB4 family type IV secretion system protein | virb4 |
| N032_RS27450 (N032_27450) | 37741..38427 | + | 687 | WP_024650164 | P-type DNA transfer protein VirB5 | - |
| N032_RS27455 (N032_27455) | 38460..38813 | + | 354 | WP_024650165 | hypothetical protein | - |
| N032_RS27460 (N032_27460) | 38824..39756 | + | 933 | WP_024650166 | type IV secretion system protein | virB6 |
| N032_RS27465 (N032_27465) | 39824..40138 | + | 315 | WP_080271125 | conjugal transfer protein | - |
| N032_RS27470 (N032_27470) | 40176..40964 | + | 789 | WP_024650167 | type IV secretion system protein | virB8 |
| N032_RS27475 (N032_27475) | 40954..41763 | + | 810 | WP_024650168 | P-type conjugative transfer protein VirB9 | virB9 |
| N032_RS27480 (N032_27480) | 41750..43099 | + | 1350 | WP_024650169 | TrbI/VirB10 family protein | virB10 |
| N032_RS27485 (N032_27485) | 43109..44164 | + | 1056 | WP_024650170 | P-type DNA transfer ATPase VirB11 | virB11 |
| N032_RS27490 (N032_27490) | 44178..44564 | + | 387 | WP_003347401 | hypothetical protein | - |
| N032_RS27495 (N032_27495) | 44549..44905 | + | 357 | WP_024650171 | hypothetical protein | - |
| N032_RS27500 (N032_27500) | 45182..45805 | + | 624 | WP_003347396 | glycoside hydrolase family 25 protein | - |
| N032_RS27510 (N032_27510) | 46140..48194 | + | 2055 | WP_032632215 | ATP-binding protein | - |
| N032_RS27515 (N032_27515) | 48196..48705 | + | 510 | WP_241197533 | hypothetical protein | - |
| N032_RS27520 (N032_27520) | 48817..49050 | + | 234 | WP_003347392 | hypothetical protein | - |
Host bacterium
| ID | 6531 | GenBank | NZ_CP034079 |
| Plasmid name | pPP1-1 | Incompatibility group | - |
| Plasmid size | 62150 bp | Coordinate of oriT [Strand] | 56510..56686 [+] |
| Host baterium | Pseudomonas syringae pv. pisi str. PP1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |