Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106064 |
| Name | oriT_X32|unnamed5 |
| Organism | Cupriavidus oxalaticus strain X32 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP038640 (64297..64395 [+], 99 nt) |
| oriT length | 99 nt |
| IRs (inverted repeats) | 24..31, 34..41 (TAACCGGC..GCCGGTTA) |
| Location of nic site | 60..61 |
| Conserved sequence flanking the nic site |
TCCTGCCCGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 99 nt
>oriT_X32|unnamed5
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
GAATAAGGGACAGTGAAGATAGATAACCGGCTCGCCGGTTAGCTAACTTCACACATCCTGCCCGCCTTACGGCGTTAATAACACCAAGGAAAGTCTACA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file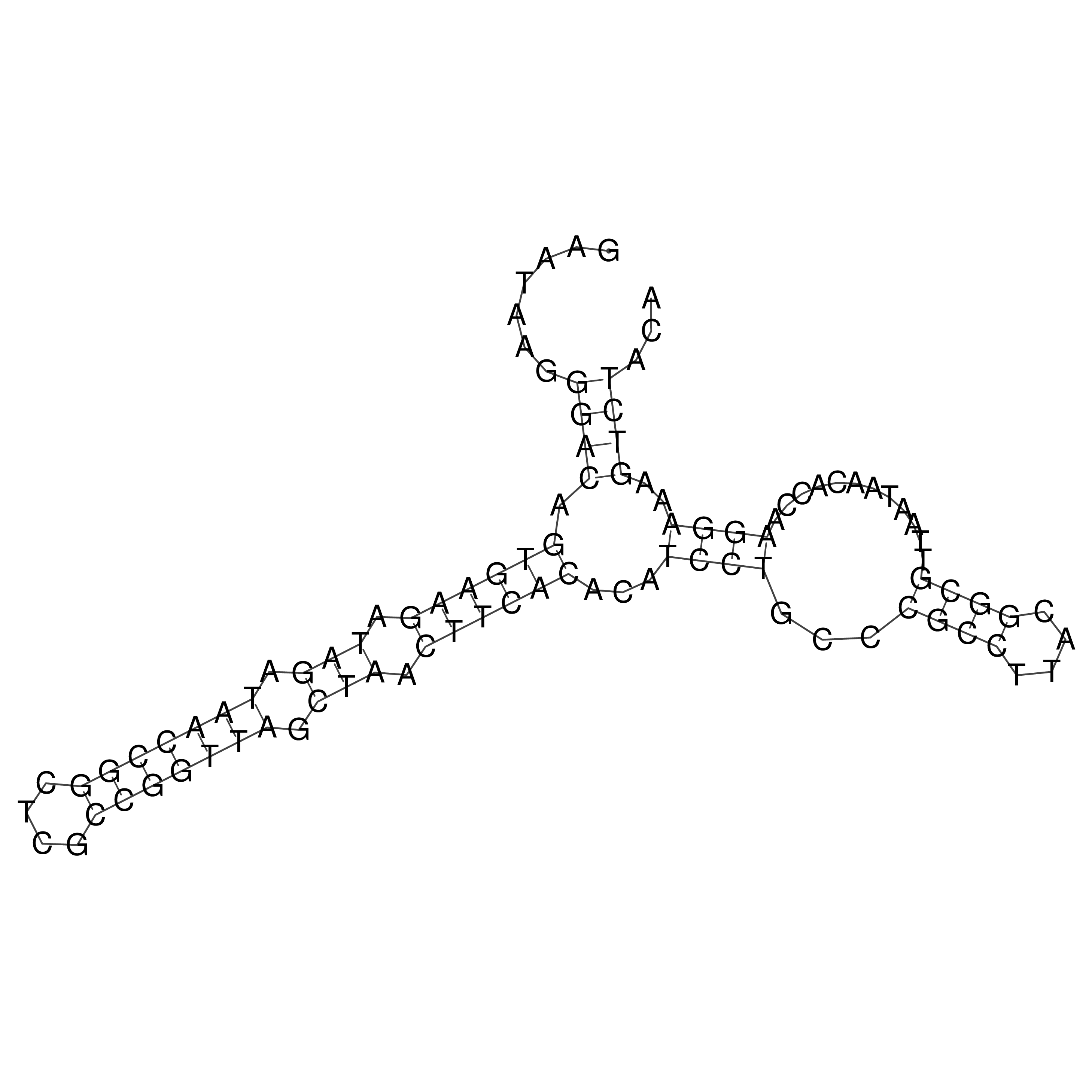
Relaxase
| ID | 4071 | GenBank | WP_011114057 |
| Name | Relaxase_E0W60_RS36755_X32|unnamed5 |
UniProt ID | A0A4P7LSA6 |
| Length | 746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 746 a.a. Molecular weight: 82038.71 Da Isoelectric Point: 10.7900
>WP_011114057.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Bacteria]
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
LQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
MIAKHVPMRSLGKSDFAGLANYITDAQSKDHRLGHVQATNCEAGSIQDAITEVLATQHTNTRAKGDKTYH
LIVSFRAGEQPSADTLRAIEERICVGLGYGEHQRISAVHNDTDNLHIHIAINKIHPTRHTMHEPYYPHRA
LAELCTALERDYGLERDNHEPRKRGAEGRAADMERHAGVESLVGWIKRECLDEIKGAQSWQELHQVMRDN
GMELRVRANGLVFEAGDGTMVKASTVARDLSKPSLEARLGPFEASPERQAQTTAKRQYRKDPIRLRVNTV
ELYAKYKAEQQSLTTARAQALERARHRKDRLIEAAKRSNRLRRATIKVVGEGRANKKLLYAQASKALRSE
LQAINKQFQQERTALYAEHSRRTWADWLKKEAQHGGADALAALRAREAAQGLKGNTIRGEGQAKPGHAPA
VDNITKKGTIIFRAGMSAVRDDGDRLQVSREATREGLQEALRLAMQRYGNRITVNGTVEFKAQMIRAAVD
SQLPITFTDPALESRRQALLNKENTHERTERPEHRGRTGRGAGGPGQRPAADQHATGAAAVARAGDGRPA
AGRGDRADAGLHAATVHRKPDVGRLGRKPPPQSQHRLRALSELGVVRIAGGSEVLLPRDVPRHVEQQGTQ
PDHALRRGISRPGTGVGQTPPGVAAADKYIAEREAKRLKGFDIPKHSRYTAGDGALTFQGTRTIEGQALA
LLKRGDEVMVMPIDQATARRLTRIAVGDAVSITAKGSIKTSKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A4P7LSA6 |
T4CP
| ID | 4085 | GenBank | WP_011114047 |
| Name | t4cp2_E0W60_RS36650_X32|unnamed5 |
UniProt ID | _ |
| Length | 852 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 852 a.a. Molecular weight: 93980.15 Da Isoelectric Point: 6.3901
>WP_011114047.1 MULTISPECIES: VirB4 family type IV secretion/conjugal transfer ATPase [Bacteria]
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
MIEAIAIAIAVLGAVLLLILFARIRAVDAELKLKKHRAKDAGLADLLNYAAVVDDGVIVGKNGSFMAAWL
YKGDDNASSTEEQREMVSFRINQALAGLGNGWMVHVDAVRRPAPNYSERGVSSFPDSVSFAIDEERRRLF
EGLGAMFEGYFVLTVTWFPPVLAQRKFVELMFDDDAVAPDHTARTTGLIAQFKREITSLESRLSSAVKMT
RLRGQKVVSEEGREVTHDDFLSWLQFCVTGLHHPVMLPSNPMYLDAVIGGQELWGGVVPKIGRKFIQVVA
IEGFPLESTPGVLSALAELPSEYRWSSRFIFMDQHESLKHLDKFRKKWKQKIRGFFDQVFNTNTGSINQD
AAAMVGDAEAAIAEVNSGLVAAGYYTSVVVLMDEDRERLAASALLVEKAVNRLAFAARIETINTLDAYLG
SLPGHGVENVRRPLINTMNLADLLPTSSIWTGSATAPCPMYPPLSPALMHCVTVGATPFRLNLHVRDLGH
TFMFGPTGAGKSTHLGIIAAQLRRYAGMSIYAFDKGMSMYPLAAGIRAATKGKSGLHFTVAADDDRLAFC
PLQFLETKGDRAWAMEWIDTILALNGVNTTPAQRNEIGNAIMSMHASGARTLSEFSVTIQDEAIREAIKQ
YTVDGSMGHLLDAEEDGLSLSDFTVFEIEELMNLGEKFALPVLLYLFRRIERALKGQPSVIILDEAWLML
GHPAFRAKIREWLKVLRKANCLVLMATQSLSDAANSGILDVIVESTATKIFLPNVYARDEDTSALYRRMG
LNARQIEILATAIPKRQYYYVSENGRRLYDLALGPMALAFVGASDKESVATIKSLEAKFGHEWVHEWLAG
RGLNLNDYLEAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4086 | GenBank | WP_000694765 |
| Name | t4cp2_E0W60_RS36750_X32|unnamed5 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69897.09 Da Isoelectric Point: 8.2993
>WP_000694765.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Bacteria]
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
MKIKMNNAVGPQVRTAKPKPSKLLPVLGAASMVGGLQAATQFFAHTFAYHATLGPNVGHVYAPWSILHWT
YKWYSQYPDEIMKAGSMGMLVSTVGLLGVAVAKVVTSNSSKANEYLHGSARWAEKKDIQAAGLLPRERNV
LEIVTGKAAPTATGVYVGGWQDKDGNFFYLRHSGPEHVLTYAPTRSGKGVGLVVPTLLSWGASSVITDLK
GELWALTAGWRQKHAKNKVLRFEPASTSGGVCWNPLDEIRLGTEYEVGDVQNLATLIVDPDGKGLDSHWQ
KTAFALLVGVILHALYKAKDDGGTATLPSVDAMLADPNRDIGELWMEMATYGHVDGQNHHAIGSAARDMM
DRPEEEAGSVLSTAKSYLALYRDPVVARNVSRSDFRIKQLMHEDDPVSLYIVTQPNDKARLRPLVRVMVN
MIVRLLADKMDFEGGRPVAHYKHRLLMMLDEFPSLGKLEIMQESLAFVAGYGIKCYLICQDINQLKSRET
GYGHDESITSNCHVQNAYPPNRVETAEHLSRLTGQTTVVKEQITTSGRRTAAMLGQVSRTYQEVQRPLLT
PDECLRMPGPKKNAQGEIEEAGDMVIYVAGYPAIYGKQPLYFKDPVFSARAAIPAPKVSDRLRAVAQAET
EGEGITI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 36750..47427
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| E0W60_RS36605 (E0W60_36605) | 32628..33437 | + | 810 | WP_005800232 | IS21-like element helper ATPase IstB | - |
| E0W60_RS37995 | 33633..33737 | - | 105 | Protein_34 | mercuric transporter MerT family protein | - |
| E0W60_RS36615 (E0W60_36615) | 33809..34243 | + | 435 | WP_003131969 | mercury resistance transcriptional regulator MerR | - |
| E0W60_RS36620 (E0W60_36620) | 34356..35576 | - | 1221 | WP_011114072 | plasmid replication initiator TrfA | - |
| E0W60_RS36625 (E0W60_36625) | 35623..35964 | - | 342 | WP_000019167 | single-stranded DNA-binding protein | - |
| E0W60_RS36630 (E0W60_36630) | 36078..36440 | + | 363 | WP_011114045 | transcriptional regulator | - |
| E0W60_RS36635 (E0W60_36635) | 36750..37712 | + | 963 | WP_000132020 | P-type conjugative transfer ATPase TrbB | virB11 |
| E0W60_RS36640 (E0W60_36640) | 37729..38193 | + | 465 | WP_011114046 | conjugal transfer system pilin TrbC | virB2 |
| E0W60_RS36645 (E0W60_36645) | 38197..38508 | + | 312 | WP_001207961 | conjugal transfer protein TrbD | virB3 |
| E0W60_RS36650 (E0W60_36650) | 38505..41063 | + | 2559 | WP_011114047 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| E0W60_RS36655 (E0W60_36655) | 41060..41842 | + | 783 | WP_011114048 | conjugal transfer protein TrbF | virB8 |
| E0W60_RS36660 (E0W60_36660) | 41860..42759 | + | 900 | WP_006122480 | P-type conjugative transfer protein TrbG | virB9 |
| E0W60_RS36665 (E0W60_36665) | 42762..43250 | + | 489 | WP_006122479 | conjugal transfer protein TrbH | - |
| E0W60_RS36670 (E0W60_36670) | 43255..44676 | + | 1422 | WP_011114050 | TrbI/VirB10 family protein | virB10 |
| E0W60_RS36675 (E0W60_36675) | 44697..45461 | + | 765 | WP_010890135 | P-type conjugative transfer protein TrbJ | virB5 |
| E0W60_RS36680 (E0W60_36680) | 45471..45698 | + | 228 | WP_000644148 | entry exclusion lipoprotein TrbK | - |
| E0W60_RS36685 (E0W60_36685) | 45709..47427 | + | 1719 | WP_011114051 | P-type conjugative transfer protein TrbL | virB6 |
| E0W60_RS36690 (E0W60_36690) | 47445..48032 | + | 588 | WP_010890138 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| E0W60_RS36695 (E0W60_36695) | 48046..48681 | + | 636 | WP_011114052 | transglycosylase SLT domain-containing protein | - |
| E0W60_RS36700 (E0W60_36700) | 48710..48976 | + | 267 | WP_010890140 | hypothetical protein | - |
| E0W60_RS36705 (E0W60_36705) | 48976..49674 | + | 699 | WP_011114053 | TraX family protein | - |
| E0W60_RS36710 (E0W60_36710) | 49690..50121 | + | 432 | WP_001665065 | OmpA family protein | - |
| E0W60_RS36715 (E0W60_36715) | 50276..50950 | + | 675 | WP_011178411 | DNA methylase | - |
| E0W60_RS36720 (E0W60_36720) | 50952..51803 | - | 852 | WP_011296189 | recombinase family protein | - |
Host bacterium
| ID | 6501 | GenBank | NZ_CP038640 |
| Plasmid name | X32|unnamed5 | Incompatibility group | IncP1 |
| Plasmid size | 89459 bp | Coordinate of oriT [Strand] | 64297..64395 [+] |
| Host baterium | Cupriavidus oxalaticus strain X32 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIF24 |