Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106060 |
| Name | oriT_pSAZ10A |
| Organism | Staphylococcus aureus strain MRSA SAZ_10 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CM016517 (6175..6210 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACG..CGTTCGC) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pSAZ10A
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file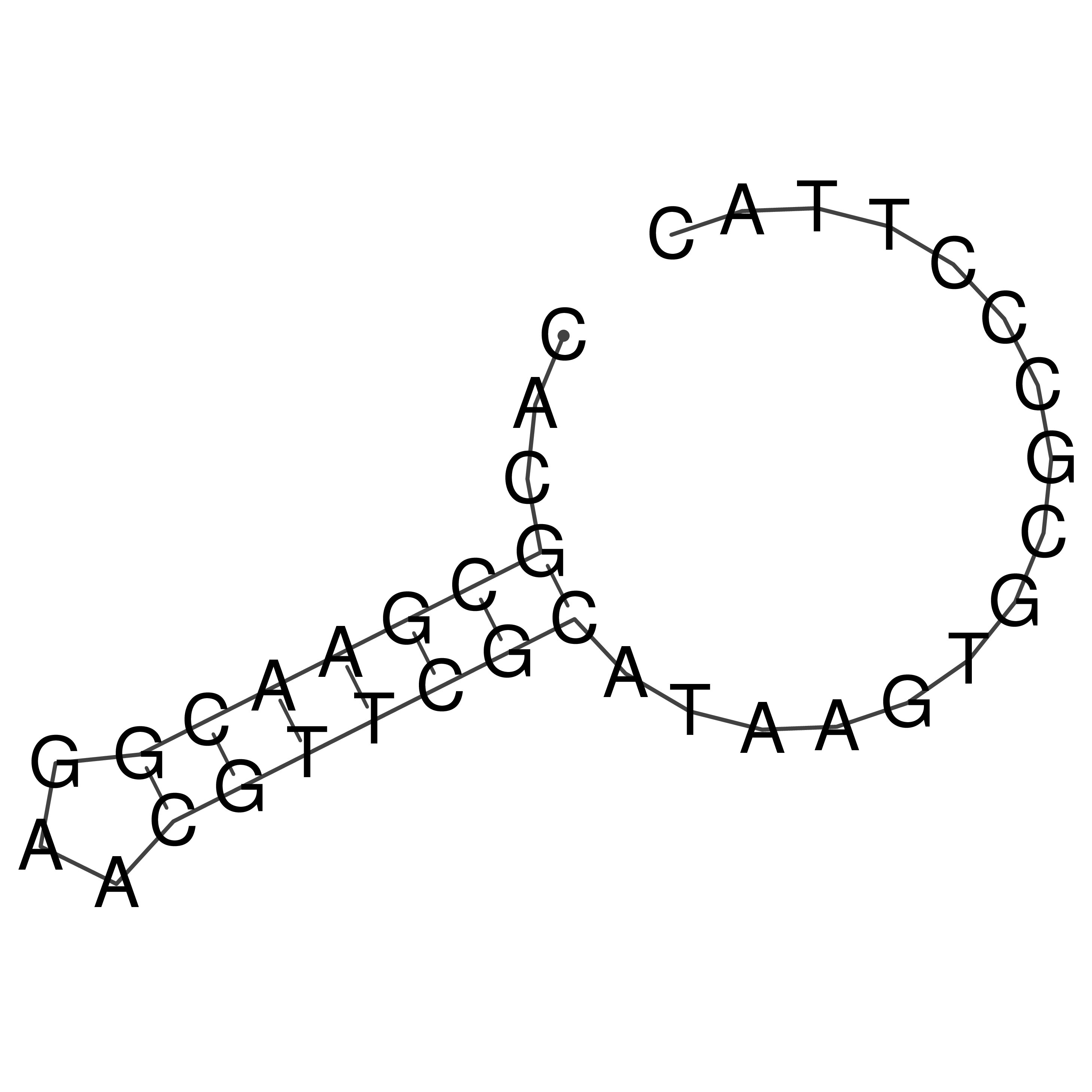
Relaxase
| ID | 4068 | GenBank | WP_001252101 |
| Name | MobA_MobL_FA040_RS00055_pSAZ10A |
UniProt ID | A0A141HMI9 |
| Length | 665 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 78908.86 Da Isoelectric Point: 5.3910
>WP_001252101.1 MULTISPECIES: MobA/MobL family protein [Bacilli]
MAMYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVHDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYKENERLYDLKQKTSLITDDEDRLNKIEDIEDRQKKLESINEVFEKQAGIFFDKNYPDQSLNYSD
DEKIFITRTILNDRDVLPANNELEDIVKEKRIKEAQISLNTVLGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHEGMEDSLKIDYYTNKLEVFRNAENILEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTIEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLSEY
DFDYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLINAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
MAMYHFQNKFVSKANGQSATAKSAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVHDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYKENERLYDLKQKTSLITDDEDRLNKIEDIEDRQKKLESINEVFEKQAGIFFDKNYPDQSLNYSD
DEKIFITRTILNDRDVLPANNELEDIVKEKRIKEAQISLNTVLGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHEGMEDSLKIDYYTNKLEVFRNAENILEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTIEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLSEY
DFDYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLINAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A141HMI9 |
T4CP
| ID | 4082 | GenBank | WP_000209436 |
| Name | t4cp2_FA040_RS00135_pSAZ10A |
UniProt ID | _ |
| Length | 546 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 546 a.a. Molecular weight: 62672.60 Da Isoelectric Point: 7.6569
>WP_000209436.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Bacilli]
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSYKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTSNIKRMQGFDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKINKDVWYKASVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNQDEEGSVELDKRFNELSKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKNKQQKLEEYKEEQ
RQKELEKEQQPQEEQQNEKVKESKDKNDQEDKKDEQQKINDSKKAFYEKLKQKQAK
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSYKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTSNIKRMQGFDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKINKDVWYKASVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNQDEEGSVELDKRFNELSKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKNKQQKLEEYKEEQ
RQKELEKEQQPQEEQQNEKVKESKDKNDQEDKKDEQQKINDSKKAFYEKLKQKQAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21981..33202
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| FA040_RS00110 (FA040_14570) | 19720..20466 | - | 747 | WP_001081004 | class I SAM-dependent methyltransferase | - |
| FA040_RS00120 (FA040_14580) | 20636..21310 | - | 675 | WP_001105984 | IS6-like element IS257 family transposase | - |
| FA040_RS16155 | 21348..21515 | - | 168 | WP_000869301 | hypothetical protein | - |
| FA040_RS00125 (FA040_14585) | 21572..21964 | - | 393 | WP_000608970 | single-stranded DNA-binding protein | - |
| FA040_RS00130 (FA040_14590) | 21981..22898 | - | 918 | WP_000383804 | conjugal transfer protein TrbL family protein | trsL |
| FA040_RS00135 (FA040_14595) | 22976..24616 | - | 1641 | WP_000209436 | VirD4-like conjugal transfer protein, CD1115 family | - |
| FA040_RS00140 (FA040_14600) | 24613..25077 | - | 465 | WP_000715273 | hypothetical protein | trsJ |
| FA040_RS00145 (FA040_14605) | 25093..27195 | - | 2103 | WP_001094109 | DNA topoisomerase III | - |
| FA040_RS00150 (FA040_14610) | 27211..27696 | - | 486 | WP_000735569 | TrsH/TraH family protein | - |
| FA040_RS00155 (FA040_14615) | 27706..28782 | - | 1077 | WP_000269385 | CHAP domain-containing protein | trsG |
| FA040_RS00160 (FA040_14620) | 28800..30080 | - | 1281 | WP_000702361 | membrane protein | trsF |
| FA040_RS00165 (FA040_14625) | 30092..32104 | - | 2013 | WP_000874919 | DUF87 domain-containing protein | virb4 |
| FA040_RS00170 (FA040_14630) | 32125..32808 | - | 684 | WP_000979860 | TrsD/TraD family conjugative transfer protein | trsD |
| FA040_RS00175 (FA040_14635) | 32795..33202 | - | 408 | WP_000110157 | hypothetical protein | trsC |
| FA040_RS00180 (FA040_14640) | 33262..33579 | - | 318 | WP_000591622 | CagC family type IV secretion system protein | - |
| FA040_RS00185 (FA040_14645) | 33596..34570 | - | 975 | WP_137359829 | conjugative transfer protein TrsA | - |
| FA040_RS00190 (FA040_14650) | 34742..34930 | + | 189 | WP_001063224 | hypothetical protein | - |
| FA040_RS00195 (FA040_14655) | 35039..35119 | - | 81 | Protein_37 | IS6 family transposase | - |
Host bacterium
| ID | 6497 | GenBank | NZ_CM016517 |
| Plasmid name | pSAZ10A | Incompatibility group | - |
| Plasmid size | 35123 bp | Coordinate of oriT [Strand] | 6175..6210 [-] |
| Host baterium | Staphylococcus aureus strain MRSA SAZ_10 |
Cargo genes
| Drug resistance gene | aadD, mupA |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |