Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106054 |
| Name | oriT_pIM007_ESBL |
| Organism | Klebsiella pneumoniae strain IM007 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP095430 (66733..66781 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pIM007_ESBL
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file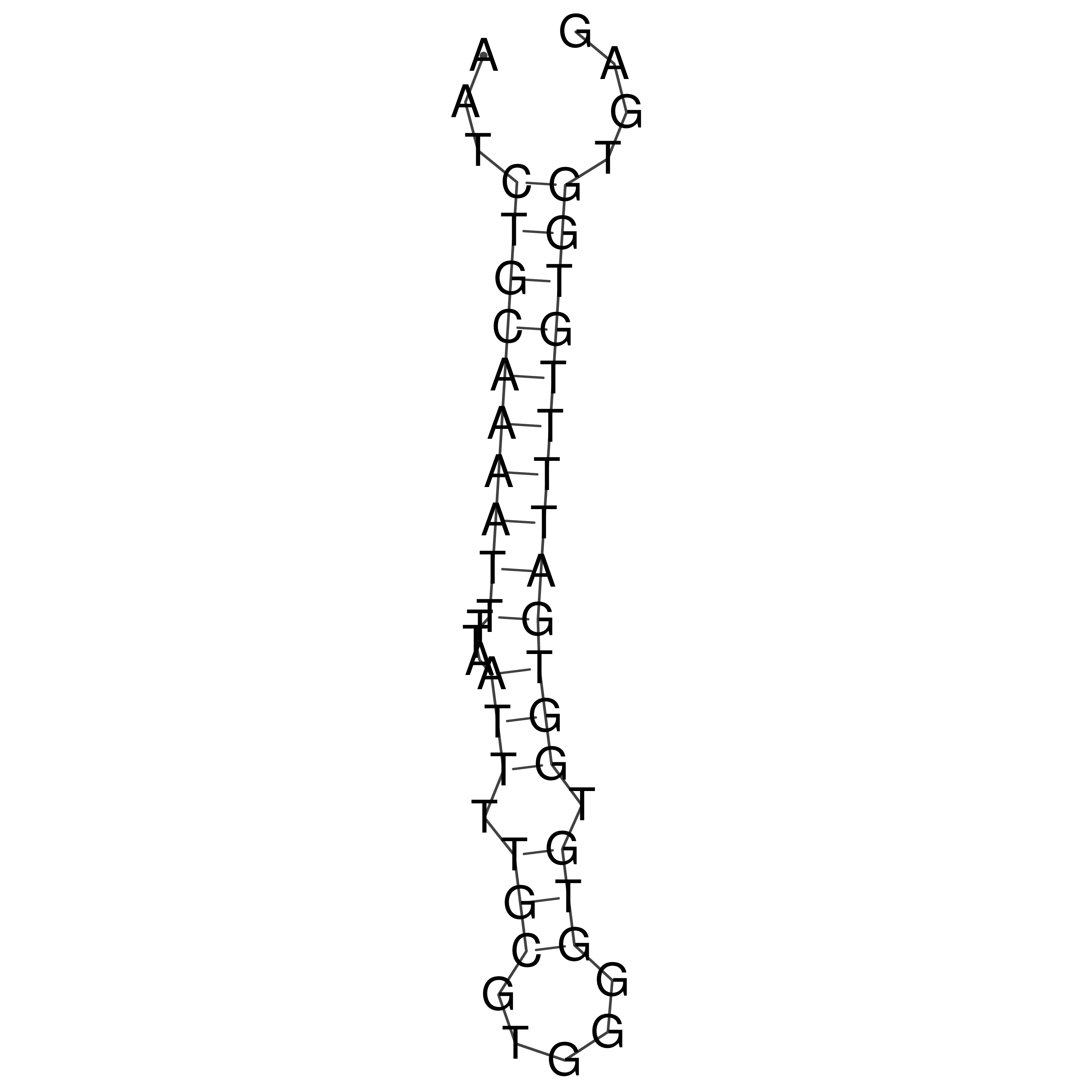
Relaxase
| ID | 4066 | GenBank | WP_047665818 |
| Name | traI_MWR71_RS29525_pIM007_ESBL |
UniProt ID | A0A0S1TL23 |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190754.90 Da Isoelectric Point: 6.3385
>WP_047665818.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKDAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDAFVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAMGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQLRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKDAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDAFVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTGSSNMLTLKDREGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAMGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQLRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLTQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKANQLITLTDSEGKERYISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKADQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKHSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4079 | GenBank | WP_013023832 |
| Name | traD_MWR71_RS29520_pIM007_ESBL |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85974.10 Da Isoelectric Point: 5.1149
>WP_013023832.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDYVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDYVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 66175..94995
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MWR71_RS29310 (MWR71_29260) | 61277..61489 | + | 213 | WP_013023815 | hypothetical protein | - |
| MWR71_RS29315 (MWR71_29265) | 61500..61724 | + | 225 | WP_004152719 | hypothetical protein | - |
| MWR71_RS29320 (MWR71_29270) | 61805..62125 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| MWR71_RS29325 (MWR71_29275) | 62115..62393 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| MWR71_RS29330 (MWR71_29280) | 62394..62807 | + | 414 | WP_013023817 | helix-turn-helix domain-containing protein | - |
| MWR71_RS29335 (MWR71_29285) | 62943..64153 | - | 1211 | Protein_87 | IS256 family transposase | - |
| MWR71_RS29340 (MWR71_29290) | 64959..65780 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| MWR71_RS29345 (MWR71_29295) | 65813..66142 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| MWR71_RS29350 (MWR71_29300) | 66175..66660 | - | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| MWR71_RS29355 (MWR71_29305) | 67092..67484 | + | 393 | WP_004194114 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| MWR71_RS29360 (MWR71_29310) | 67714..68415 | + | 702 | WP_004194113 | hypothetical protein | - |
| MWR71_RS29365 (MWR71_29315) | 68501..68701 | + | 201 | WP_004194116 | TraY domain-containing protein | - |
| MWR71_RS29370 (MWR71_29320) | 68770..69138 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| MWR71_RS29375 (MWR71_29325) | 69152..69457 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| MWR71_RS29380 (MWR71_29330) | 69477..70043 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| MWR71_RS29385 (MWR71_29335) | 70030..70770 | + | 741 | WP_013023821 | type-F conjugative transfer system secretin TraK | traK |
| MWR71_RS29390 (MWR71_29340) | 70770..72194 | + | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| MWR71_RS29395 (MWR71_29345) | 72308..72892 | + | 585 | WP_013023822 | type IV conjugative transfer system lipoprotein TraV | traV |
| MWR71_RS29400 (MWR71_29350) | 73024..73434 | + | 411 | WP_009309869 | hypothetical protein | - |
| MWR71_RS29405 (MWR71_29355) | 73540..73758 | + | 219 | WP_015344987 | hypothetical protein | - |
| MWR71_RS29410 (MWR71_29360) | 73759..74070 | + | 312 | WP_015344986 | hypothetical protein | - |
| MWR71_RS29415 (MWR71_29365) | 74137..74541 | + | 405 | WP_004197817 | hypothetical protein | - |
| MWR71_RS29420 (MWR71_29370) | 74917..75315 | + | 399 | WP_013609531 | hypothetical protein | - |
| MWR71_RS29425 (MWR71_29375) | 75387..78026 | + | 2640 | WP_013023824 | type IV secretion system protein TraC | virb4 |
| MWR71_RS29430 (MWR71_29380) | 78026..78415 | + | 390 | WP_004197815 | type-F conjugative transfer system protein TrbI | - |
| MWR71_RS29435 (MWR71_29385) | 78415..79041 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| MWR71_RS29440 (MWR71_29390) | 79083..79472 | + | 390 | WP_004194992 | hypothetical protein | - |
| MWR71_RS29445 (MWR71_29395) | 79469..80458 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| MWR71_RS29450 (MWR71_29400) | 80471..81109 | + | 639 | WP_011977786 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| MWR71_RS29455 (MWR71_29405) | 81168..83123 | + | 1956 | WP_013023827 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| MWR71_RS29460 (MWR71_29410) | 83155..83409 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| MWR71_RS29465 (MWR71_29415) | 83387..83635 | + | 249 | WP_004152675 | hypothetical protein | - |
| MWR71_RS29470 (MWR71_29420) | 83648..83974 | + | 327 | WP_004152676 | hypothetical protein | - |
| MWR71_RS29475 (MWR71_29425) | 83995..84747 | + | 753 | WP_048337413 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| MWR71_RS29480 (MWR71_29430) | 84758..84997 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| MWR71_RS29485 (MWR71_29435) | 84969..85526 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| MWR71_RS29490 (MWR71_29440) | 85572..86015 | + | 444 | WP_013023828 | F-type conjugal transfer protein TrbF | - |
| MWR71_RS29495 (MWR71_29445) | 85993..87372 | + | 1380 | WP_014343487 | conjugal transfer pilus assembly protein TraH | traH |
| MWR71_RS29500 (MWR71_29450) | 87372..90221 | + | 2850 | WP_013609534 | conjugal transfer mating-pair stabilization protein TraG | traG |
| MWR71_RS29505 (MWR71_29455) | 90224..90757 | + | 534 | WP_014343486 | conjugal transfer protein TraS | - |
| MWR71_RS29510 (MWR71_29460) | 90943..91674 | + | 732 | WP_013023830 | conjugal transfer complement resistance protein TraT | - |
| MWR71_RS29515 (MWR71_29465) | 91867..92556 | + | 690 | WP_013023831 | hypothetical protein | - |
| MWR71_RS29520 (MWR71_29470) | 92683..94995 | + | 2313 | WP_013023832 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 6491 | GenBank | NZ_CP095430 |
| Plasmid name | pIM007_ESBL | Incompatibility group | IncFII |
| Plasmid size | 124551 bp | Coordinate of oriT [Strand] | 66733..66781 [-] |
| Host baterium | Klebsiella pneumoniae strain IM007 |
Cargo genes
| Drug resistance gene | mph(A), tet(A), floR, qnrS1, blaCTX-M-3, blaTEM-1B, aac(6')-Ib-cr, ARR-3, dfrA27, aadA16, sul1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |