Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 106029 |
| Name | oriT_pRes_C2582 |
| Organism | Klebsiella pneumoniae strain C2582 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP079210 (78589..78712 [+], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 101..106, 119..124 (TTTAAT..ATTAAA) 91..99, 113..121 (AATAATGTA..TACATTATT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
>oriT_pRes_C2582
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
GGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTGCTTTTTGAATCATTAGCTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATTAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file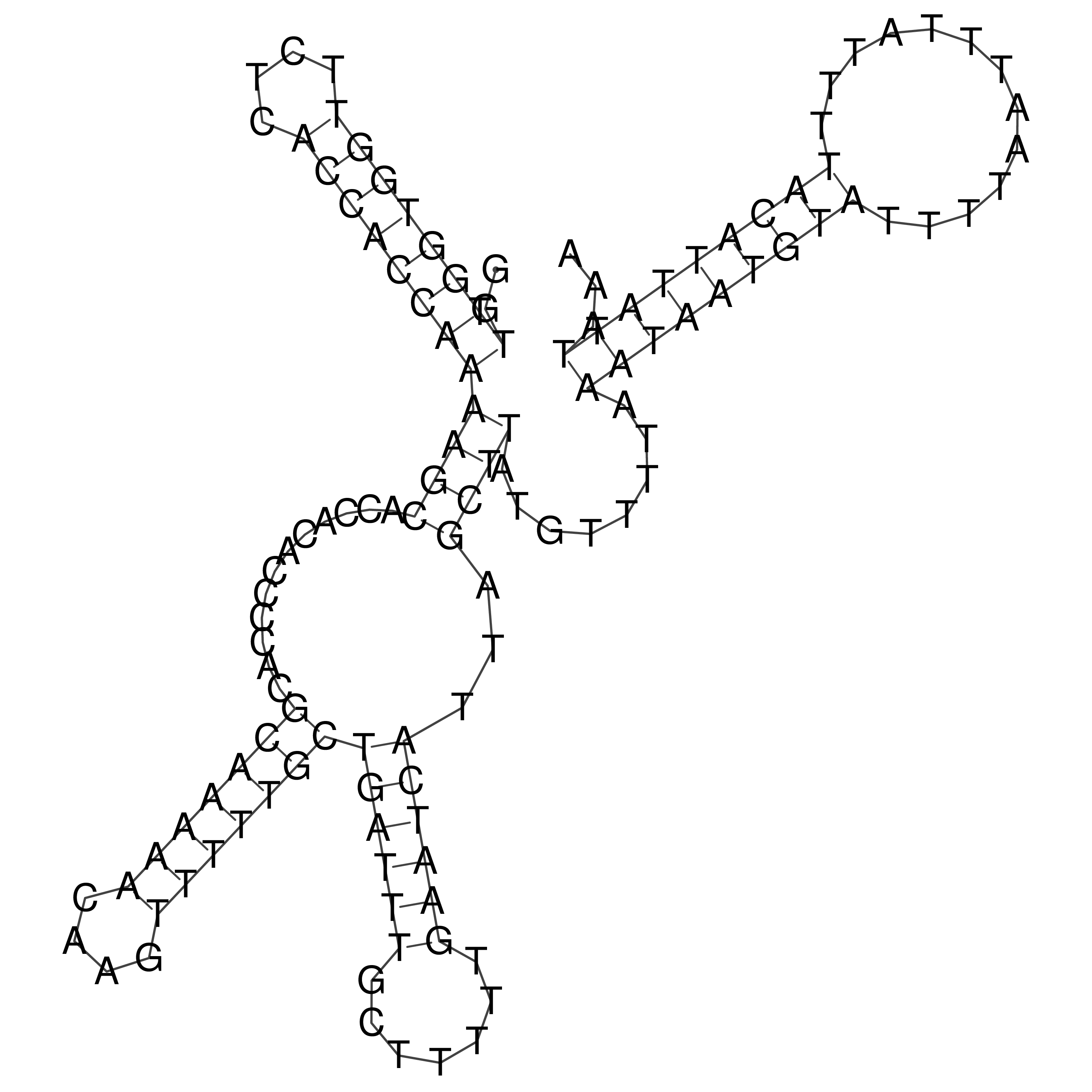
T4CP
| ID | 4070 | GenBank | WP_015059012 |
| Name | traC_KV145_RS29075_pRes_C2582 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99393.22 Da Isoelectric Point: 6.3465
>WP_015059012.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAAA
TQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKKQARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAAA
TQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKKQARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 78017..90660
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| KV145_RS28970 (KV145_28875) | 73989..74423 | + | 435 | WP_000845953 | conjugation system SOS inhibitor PsiB | - |
| KV145_RS28975 (KV145_28880) | 74420..75139 | + | 720 | WP_001276217 | plasmid SOS inhibition protein A | - |
| KV145_RS28980 | 75361..75510 | + | 150 | Protein_108 | plasmid maintenance protein Mok | - |
| KV145_RS28985 (KV145_28885) | 75452..75577 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| KV145_RS28990 (KV145_28890) | 75896..76192 | - | 297 | Protein_110 | hypothetical protein | - |
| KV145_RS28995 (KV145_28895) | 76492..76788 | + | 297 | WP_001272251 | hypothetical protein | - |
| KV145_RS29000 (KV145_28900) | 76899..77720 | + | 822 | WP_001234445 | DUF932 domain-containing protein | - |
| KV145_RS29005 (KV145_28905) | 78017..78664 | - | 648 | WP_015059008 | transglycosylase SLT domain-containing protein | virB1 |
| KV145_RS29010 (KV145_28910) | 78941..79324 | + | 384 | WP_000124981 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| KV145_RS29015 (KV145_28915) | 79515..80201 | + | 687 | WP_015059009 | PAS domain-containing protein | - |
| KV145_RS29020 (KV145_28920) | 80295..80522 | + | 228 | WP_001254386 | conjugal transfer relaxosome protein TraY | - |
| KV145_RS29025 (KV145_28925) | 80556..80921 | + | 366 | WP_021519752 | type IV conjugative transfer system pilin TraA | - |
| KV145_RS29030 (KV145_28930) | 80936..81247 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| KV145_RS29035 (KV145_28935) | 81269..81835 | + | 567 | WP_000399794 | type IV conjugative transfer system protein TraE | traE |
| KV145_RS29040 (KV145_28940) | 81822..82550 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| KV145_RS29045 (KV145_28945) | 82550..83977 | + | 1428 | WP_032297072 | F-type conjugal transfer pilus assembly protein TraB | traB |
| KV145_RS29050 (KV145_28950) | 83967..84557 | + | 591 | WP_000002778 | conjugal transfer pilus-stabilizing protein TraP | - |
| KV145_RS29055 (KV145_28955) | 84544..84741 | + | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| KV145_RS29060 (KV145_28960) | 84753..85004 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| KV145_RS29065 (KV145_28965) | 85001..85516 | + | 516 | WP_062955122 | type IV conjugative transfer system lipoprotein TraV | traV |
| KV145_RS29070 (KV145_28970) | 85651..85872 | + | 222 | WP_001278692 | conjugal transfer protein TraR | - |
| KV145_RS29075 (KV145_28975) | 86032..88659 | + | 2628 | WP_015059012 | type IV secretion system protein TraC | virb4 |
| KV145_RS29080 (KV145_28980) | 88656..89042 | + | 387 | WP_015059013 | type-F conjugative transfer system protein TrbI | - |
| KV145_RS29085 (KV145_28985) | 89039..89671 | + | 633 | WP_001203735 | type-F conjugative transfer system protein TraW | traW |
| KV145_RS29090 (KV145_28990) | 89668..90660 | + | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| KV145_RS29095 (KV145_28995) | 90687..90995 | + | 309 | WP_000412447 | hypothetical protein | - |
| KV145_RS29100 (KV145_29000) | 91058..91579 | + | 522 | WP_015059014 | hypothetical protein | - |
| KV145_RS29105 (KV145_29005) | 91606..92121 | + | 516 | Protein_133 | type-F conjugative transfer system pilin assembly protein TrbC | - |
| KV145_RS29110 (KV145_29010) | 92183..92887 | + | 705 | WP_001067855 | IS6-like element IS26 family transposase | - |
| KV145_RS29115 (KV145_29015) | 92833..93264 | - | 432 | WP_011977770 | class II aldolase/adducin family protein | - |
| KV145_RS29120 (KV145_29020) | 93257..94509 | - | 1253 | Protein_136 | 3-oxo-tetronate kinase | - |
| KV145_RS29125 (KV145_29025) | 94521..95423 | - | 903 | WP_002903955 | L-threonate dehydrogenase | - |
Host bacterium
| ID | 6466 | GenBank | NZ_CP079210 |
| Plasmid name | pRes_C2582 | Incompatibility group | IncFII |
| Plasmid size | 128399 bp | Coordinate of oriT [Strand] | 78589..78712 [+] |
| Host baterium | Klebsiella pneumoniae strain C2582 |
Cargo genes
| Drug resistance gene | blaCTX-M-65, fosA3, blaTEM-1B, rmtB, blaSHV-12, blaKPC-2 |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |