Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105997 |
| Name | oriT_pKpn13-1 |
| Organism | Klebsiella pneumoniae strain Kpn-13 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP047708 (143513..143562 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pKpn13-1
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file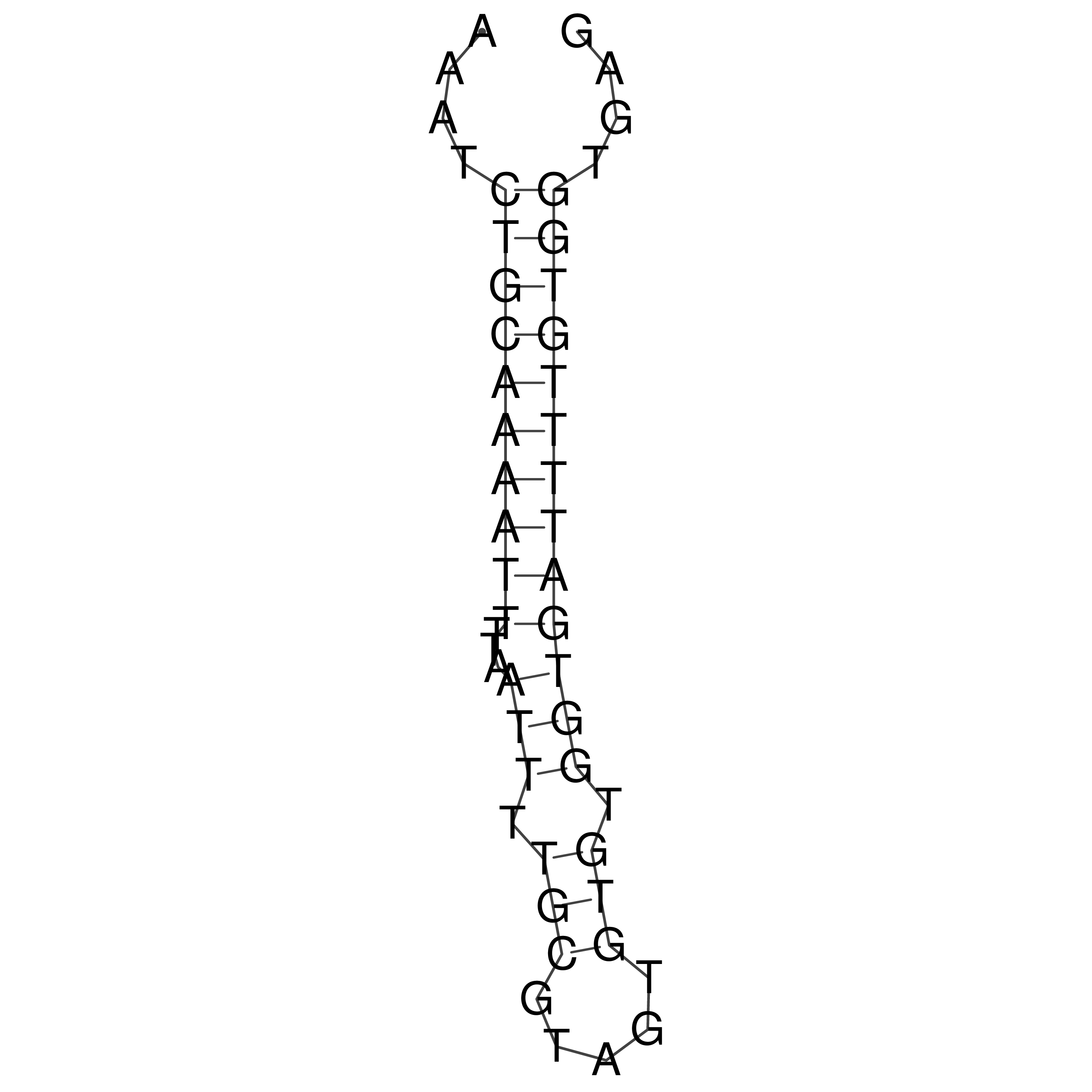
Relaxase
| ID | 4032 | GenBank | WP_023287123 |
| Name | traI_GVI78_RS25750_pKpn13-1 |
UniProt ID | A0A486VBD3 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190201.88 Da Isoelectric Point: 6.2218
>WP_023287123.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINTTQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARAGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGAREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTIQ
AQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPAQEKAIQK
ALSEGETLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINTTQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARAGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGAREQSVLNGLIRDSLRHEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTIQ
AQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPAQEKAIQK
ALSEGETLPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVSGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAITGGRVWADPAPVGPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4044 | GenBank | WP_023313941 |
| Name | traD_GVI78_RS25755_pKpn13-1 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85666.78 Da Isoelectric Point: 5.2499
>WP_023313941.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGTADTNSHAGEQPEPVSQPA
PAEVTVSPAPVKAPATTKMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNKDGVIEDMQAYDAWADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGTADTNSHAGEQPEPVSQPA
PAEVTVSPAPVKAPATTKMPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNKDGVIEDMQAYDAWADEQTLRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 122484..144114
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GVI78_RS25755 (GVI78_25415) | 122484..124793 | - | 2310 | WP_023313941 | type IV conjugative transfer system coupling protein TraD | virb4 |
| GVI78_RS25760 | 124920..125609 | - | 690 | WP_013023831 | hypothetical protein | - |
| GVI78_RS25765 (GVI78_25425) | 125802..126236 | - | 435 | Protein_122 | complement resistance protein TraT | - |
| GVI78_RS25770 (GVI78_25430) | 126251..127219 | + | 969 | WP_015958259 | IS5 family transposase | - |
| GVI78_RS25775 (GVI78_25435) | 127278..128921 | - | 1644 | Protein_124 | conjugal transfer protein TraN | - |
| GVI78_RS25780 (GVI78_25440) | 128980..129618 | - | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| GVI78_RS25785 (GVI78_25445) | 129631..130620 | - | 990 | WP_013214027 | conjugal transfer pilus assembly protein TraU | traU |
| GVI78_RS25790 (GVI78_25450) | 130617..131018 | - | 402 | WP_004194979 | hypothetical protein | - |
| GVI78_RS25795 (GVI78_25455) | 131053..131688 | - | 636 | WP_013214026 | type-F conjugative transfer system protein TraW | traW |
| GVI78_RS25800 (GVI78_25460) | 131688..132077 | - | 390 | WP_013214025 | type-F conjugative transfer system protein TrbI | - |
| GVI78_RS25805 (GVI78_25465) | 132077..134716 | - | 2640 | WP_032431388 | type IV secretion system protein TraC | virb4 |
| GVI78_RS25810 (GVI78_25470) | 134788..135186 | - | 399 | WP_023179972 | hypothetical protein | - |
| GVI78_RS25815 (GVI78_25475) | 135565..135969 | - | 405 | WP_197384183 | hypothetical protein | - |
| GVI78_RS25820 (GVI78_25480) | 136036..136347 | - | 312 | WP_197384184 | hypothetical protein | - |
| GVI78_RS25825 (GVI78_25485) | 136348..136566 | - | 219 | WP_004195468 | hypothetical protein | - |
| GVI78_RS25830 (GVI78_25490) | 136889..137473 | - | 585 | WP_197384185 | type IV conjugative transfer system lipoprotein TraV | traV |
| GVI78_RS25835 (GVI78_25495) | 137496..138092 | - | 597 | WP_197384186 | conjugal transfer pilus-stabilizing protein TraP | - |
| GVI78_RS25840 (GVI78_25500) | 138085..139509 | - | 1425 | WP_004233801 | F-type conjugal transfer pilus assembly protein TraB | traB |
| GVI78_RS25845 (GVI78_25505) | 139509..140249 | - | 741 | WP_197384187 | type-F conjugative transfer system secretin TraK | traK |
| GVI78_RS25850 (GVI78_25510) | 140236..140802 | - | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| GVI78_RS25855 (GVI78_25515) | 140822..141127 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| GVI78_RS25860 (GVI78_25520) | 141141..141509 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| GVI78_RS25865 (GVI78_25525) | 141571..141777 | - | 207 | WP_171773970 | TraY domain-containing protein | - |
| GVI78_RS25870 (GVI78_25530) | 141910..142629 | - | 720 | WP_197384188 | conjugal transfer protein TrbJ | - |
| GVI78_RS25875 (GVI78_25535) | 142828..143244 | - | 417 | WP_167878886 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| GVI78_RS25880 (GVI78_25540) | 143707..144114 | + | 408 | WP_225011827 | transglycosylase SLT domain-containing protein | virB1 |
| GVI78_RS25885 (GVI78_25545) | 144152..144682 | - | 531 | WP_049070338 | antirestriction protein | - |
| GVI78_RS25890 (GVI78_25550) | 144874..145104 | + | 231 | WP_197384202 | hypothetical protein | - |
| GVI78_RS25895 (GVI78_25555) | 145199..145903 | - | 705 | WP_117048525 | IS6-like element IS26 family transposase | - |
| GVI78_RS25900 (GVI78_25560) | 145980..146906 | - | 927 | WP_001554380 | LysR family transcriptional regulator | - |
| GVI78_RS25905 (GVI78_25565) | 147036..147824 | + | 789 | WP_001554379 | alpha/beta hydrolase | - |
| GVI78_RS25910 (GVI78_25570) | 148016..148195 | + | 180 | Protein_151 | hypothetical protein | - |
Host bacterium
| ID | 6434 | GenBank | NZ_CP047708 |
| Plasmid name | pKpn13-1 | Incompatibility group | IncFIB |
| Plasmid size | 181030 bp | Coordinate of oriT [Strand] | 143513..143562 [+] |
| Host baterium | Klebsiella pneumoniae strain Kpn-13 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoS, pcoE, arsH, arsC, arsB, arsA, arsD, arsR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |