Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105955 |
| Name | oriT_NER9|pB |
| Organism | Ensifer adhaerens strain NER9 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP101519 (1305751..1305794 [-], 44 nt) |
| oriT length | 44 nt |
| IRs (inverted repeats) | 23..28, 33..38 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 44 nt
>oriT_NER9|pB
GGATCCGAAGGGCGCAATTATACGTCGCTGACGCGACGCTTTGC
GGATCCGAAGGGCGCAATTATACGTCGCTGACGCGACGCTTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file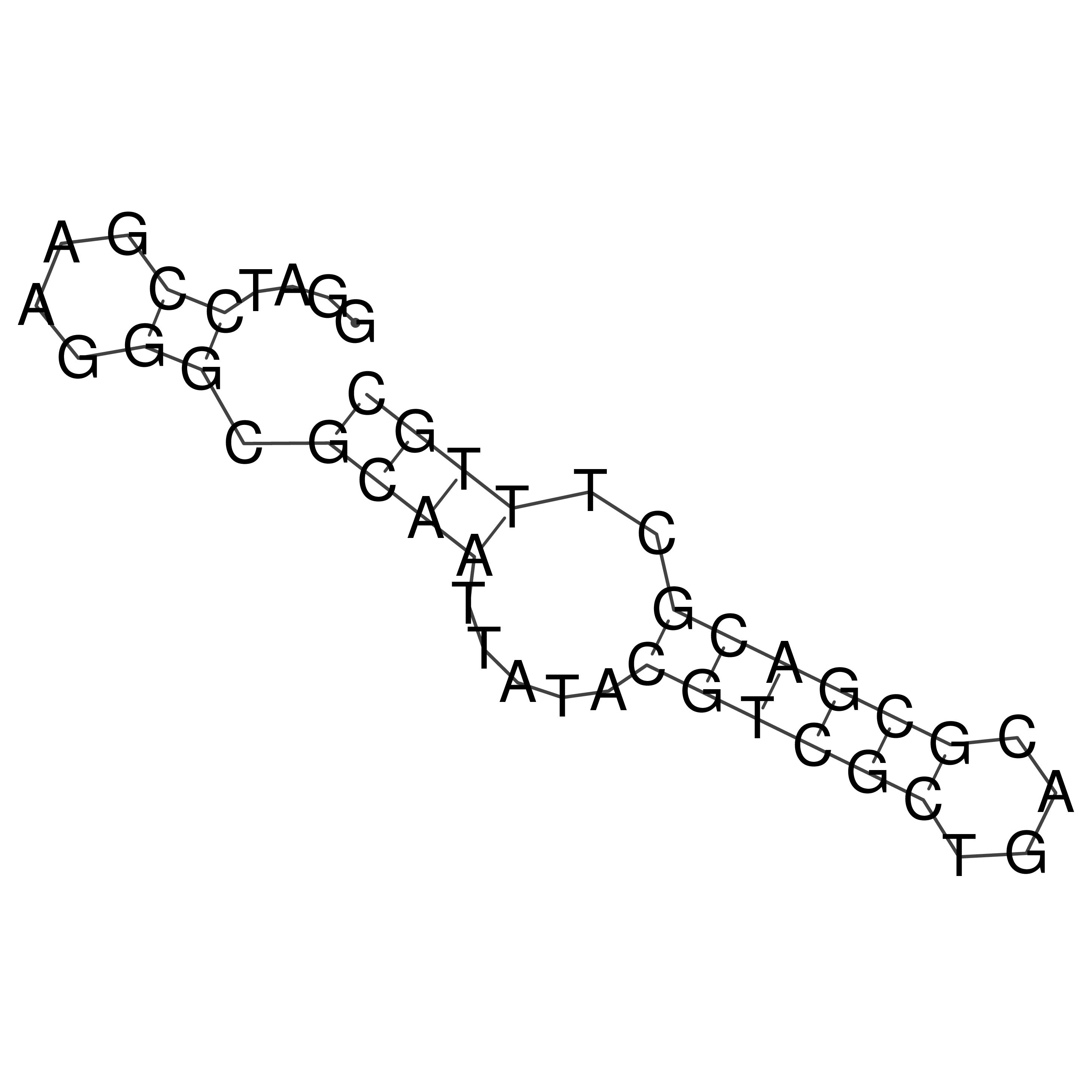
Relaxase
| ID | 4012 | GenBank | WP_255381715 |
| Name | traA_MYG64_RS34030_NER9|pB |
UniProt ID | _ |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 121714.85 Da Isoelectric Point: 9.8338
>WP_255381715.1 Ti-type conjugative transfer relaxase TraA [Ensifer adhaerens]
MAAPHFSVSIVARGSGRSAVLSAAYRHCAKMDYKREARTIDYTRKQGLLHEEFVIPADAPEWLQSMIAYH
SVSGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELSAAQNIELVRDFVARHITGQGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGPDGKPLRNDAGKIVYQLWAGSLNDFIVFRDGWFACQNRHLALAG
LDLRVDGRSFEKQGIALMPAIHLGVGAKAMERKATGCGEVAALERLELQAERRAENARRIQRNPGIVLDL
LTREKSVFDSQDIAKILHRYIDDAGHFQNLMARILESPETLRLNLERIDFATGLRAPAKYTTRELIRLEA
GMANRAIWLSQRSSHEVRRPVLDATFSRHDRLSDEQKTAIEHVVGKARIALVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKATEGLEKEAGIASRTLASWEMCWGKGREQLDDRCVFVLDEAGMVSSRQMALFVE
AVTSCGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELEIIYRQREQWMRDASLDLARGNVGKAVEAYR
ANGRMIGAELKSEAVAALIADWNRDFDPAKSSLILAHLRRDVRMLNELARAKLIQRGIIESGATFKTADG
DRSFAAGDQIVFLKNEGSLGVKNGMLAKVADAAPGRIVAEIGDGKHRRQISVDQRFYSNVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGRRSFAKAGGLIAILSRKNSKETTLDYVNGSFY
RQALLFAEARGFHLVNVARTLARDRLVWVVRQKQRLVELGAQLSALGQALGLSPIKKSSETASAINEAKP
MVSGIKVFPKSLEQTVEDRLAADPGLKKQWEEVSSRFHVVYAEPEAAFKAINVDAILADEAAAKSAIAKI
NTEPESFGALKGKTGMFVSSAHKQDREKATLNAPALARNLERYLRHRALAERKHETEERSIRAKFSIDIP
ALSPSAKQTLERVRDAIDRNDLPAGLEFALAEGQVKAELDGFTRAVSERFGERTFLTLTAKEPNGETFQR
LTSDMNPSQKGEVQSAWNAMRTAQQLAAHEHTMTALKQSETSRQTKSQGLSLK
MAAPHFSVSIVARGSGRSAVLSAAYRHCAKMDYKREARTIDYTRKQGLLHEEFVIPADAPEWLQSMIAYH
SVSGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELSAAQNIELVRDFVARHITGQGMVADWVFHDAPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGPDGKPLRNDAGKIVYQLWAGSLNDFIVFRDGWFACQNRHLALAG
LDLRVDGRSFEKQGIALMPAIHLGVGAKAMERKATGCGEVAALERLELQAERRAENARRIQRNPGIVLDL
LTREKSVFDSQDIAKILHRYIDDAGHFQNLMARILESPETLRLNLERIDFATGLRAPAKYTTRELIRLEA
GMANRAIWLSQRSSHEVRRPVLDATFSRHDRLSDEQKTAIEHVVGKARIALVIGRAGAGKTTMMKAAREA
WEAAGYRVVGGALAGKATEGLEKEAGIASRTLASWEMCWGKGREQLDDRCVFVLDEAGMVSSRQMALFVE
AVTSCGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELEIIYRQREQWMRDASLDLARGNVGKAVEAYR
ANGRMIGAELKSEAVAALIADWNRDFDPAKSSLILAHLRRDVRMLNELARAKLIQRGIIESGATFKTADG
DRSFAAGDQIVFLKNEGSLGVKNGMLAKVADAAPGRIVAEIGDGKHRRQISVDQRFYSNVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLGVYYGRRSFAKAGGLIAILSRKNSKETTLDYVNGSFY
RQALLFAEARGFHLVNVARTLARDRLVWVVRQKQRLVELGAQLSALGQALGLSPIKKSSETASAINEAKP
MVSGIKVFPKSLEQTVEDRLAADPGLKKQWEEVSSRFHVVYAEPEAAFKAINVDAILADEAAAKSAIAKI
NTEPESFGALKGKTGMFVSSAHKQDREKATLNAPALARNLERYLRHRALAERKHETEERSIRAKFSIDIP
ALSPSAKQTLERVRDAIDRNDLPAGLEFALAEGQVKAELDGFTRAVSERFGERTFLTLTAKEPNGETFQR
LTSDMNPSQKGEVQSAWNAMRTAQQLAAHEHTMTALKQSETSRQTKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4021 | GenBank | WP_112984549 |
| Name | t4cp2_MYG64_RS33970_NER9|pB |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91721.71 Da Isoelectric Point: 6.6908
>WP_112984549.1 conjugal transfer protein TrbE [Ensifer adhaerens]
MVALKQFRAKGPSFSDLVPYAGMVDNGVLLLKDGSLMSAWYFAGPDLESATDQERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSQERCHFPDPVTRAIDAERRMHFARDNGHFESKHALVLTYRPPESRKAVLSK
YIYSDEKSRKKSYADTVLIAFKNAVREVEQYFANTLSLHRMRTRAIADRGGARTAHYDELLQFARFCITG
ENHPIRLPDVPMYLDWIATAELQHGLTPLVENRFLGVVAIDGFPAESWPGILNSLDLMPLAYRWSSRFVF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDALTMVAETEDAIAQASSQLVAYGYYTTVVVL
FDGNKEALQEKAEAVRRLVQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTNNLADLIPLNSVWS
GDPVAPSPFYPQNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGNSLLPLTLAAGGDHYEIGSDEKGRALAFCPLSELSTDEDRAWATEWIEMLVALQGVTITPDLRNAI
ARQIRLMASAPGRSLSDFVSGVQLREIKDALHHYTVDGPMGHLLDAEHDGLSLRSLQCFEIEQLMNMGER
NLVPVLTYLFRRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERSG
IIDVLKESCLTKICLPNGAAREPGTRAFYERIGFNERQIEIVSMATPKREYYVVTPEGRRLFDMALGPFA
LSFVGASGKEDLKRIRALRSEHGRDWPTQWLKTRGVQDAASLLNIE
MVALKQFRAKGPSFSDLVPYAGMVDNGVLLLKDGSLMSAWYFAGPDLESATDQERNELSRQINAVLSRLG
SGWMIQVEAIRIPTVDYPSQERCHFPDPVTRAIDAERRMHFARDNGHFESKHALVLTYRPPESRKAVLSK
YIYSDEKSRKKSYADTVLIAFKNAVREVEQYFANTLSLHRMRTRAIADRGGARTAHYDELLQFARFCITG
ENHPIRLPDVPMYLDWIATAELQHGLTPLVENRFLGVVAIDGFPAESWPGILNSLDLMPLAYRWSSRFVF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDALTMVAETEDAIAQASSQLVAYGYYTTVVVL
FDGNKEALQEKAEAVRRLVQAEGFGARIETLNATDAYLGSLPGNWYCNIREPLINTNNLADLIPLNSVWS
GDPVAPSPFYPQNSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEHAQIFA
FDKGNSLLPLTLAAGGDHYEIGSDEKGRALAFCPLSELSTDEDRAWATEWIEMLVALQGVTITPDLRNAI
ARQIRLMASAPGRSLSDFVSGVQLREIKDALHHYTVDGPMGHLLDAEHDGLSLRSLQCFEIEQLMNMGER
NLVPVLTYLFRRIEKRLDGSPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERSG
IIDVLKESCLTKICLPNGAAREPGTRAFYERIGFNERQIEIVSMATPKREYYVVTPEGRRLFDMALGPFA
LSFVGASGKEDLKRIRALRSEHGRDWPTQWLKTRGVQDAASLLNIE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4022 | GenBank | WP_112975486 |
| Name | traG_MYG64_RS34045_NER9|pB |
UniProt ID | _ |
| Length | 638 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 638 a.a. Molecular weight: 68662.37 Da Isoelectric Point: 9.1933
>WP_112975486.1 Ti-type conjugative transfer system protein TraG [Ensifer adhaerens]
MTATRTLLVVAPVALMVTAIFVTTGMETWLQGFGNTEAAKVALGRVGISAPYIAASAVGVVFLFASAGSA
SVRFSGWGVVAGSGATILIAALREAVRLAAFAGKMPAGKNLAASLDPATVLGAAIALMAASFALRVVLIG
NAAFAHPEPKRVNGRRALHGEADWMKLASAARLFPESGGIVVGELYRVDKDSTAAQSFRADSPDTWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSVVILDPSKEVAPMVSKHRRDADRDVFVLD
PMAPETGFNALDWIGRFGGTKEEDIASVASWIMSESGGVRGVRDDFFRASALQLLTALIADVCLSGRTDK
ADQTLRQVRKNLSEPEPKLRERLQTIYDNSDSDFVKENVAAFVNMTPETFSGVYANAVRETHWLSYSNYA
ALVSGSTFSTGEIATGHADVFINVDLKTLEGHSGLARVIIGALLNAIYNRNGEMNGRTLFLLDEVSRLGY
MRVLETARDAGRKYGITLTMIYQSIGQMREAYGGRDAASKWFESVSWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQSRGSSRTRSKQLAARPLIQPHEVLQMRADEQIVFTAGNPPLRCGRAIWFRRADMKSCVA
ENRFERMK
MTATRTLLVVAPVALMVTAIFVTTGMETWLQGFGNTEAAKVALGRVGISAPYIAASAVGVVFLFASAGSA
SVRFSGWGVVAGSGATILIAALREAVRLAAFAGKMPAGKNLAASLDPATVLGAAIALMAASFALRVVLIG
NAAFAHPEPKRVNGRRALHGEADWMKLASAARLFPESGGIVVGELYRVDKDSTAAQSFRADSPDTWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSVVILDPSKEVAPMVSKHRRDADRDVFVLD
PMAPETGFNALDWIGRFGGTKEEDIASVASWIMSESGGVRGVRDDFFRASALQLLTALIADVCLSGRTDK
ADQTLRQVRKNLSEPEPKLRERLQTIYDNSDSDFVKENVAAFVNMTPETFSGVYANAVRETHWLSYSNYA
ALVSGSTFSTGEIATGHADVFINVDLKTLEGHSGLARVIIGALLNAIYNRNGEMNGRTLFLLDEVSRLGY
MRVLETARDAGRKYGITLTMIYQSIGQMREAYGGRDAASKWFESVSWISFAAINDPETADYISRRCGMTT
VEIDQVSRSFQSRGSSRTRSKQLAARPLIQPHEVLQMRADEQIVFTAGNPPLRCGRAIWFRRADMKSCVA
ENRFERMK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 1288958..1300634
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MYG64_RS33935 (MYG64_33795) | 1284579..1285277 | + | 699 | WP_089045403 | LuxR family transcriptional regulator | - |
| MYG64_RS33940 (MYG64_33800) | 1285614..1286861 | - | 1248 | WP_255381701 | HipA domain-containing protein | - |
| MYG64_RS33945 (MYG64_33805) | 1286854..1287189 | - | 336 | WP_029742477 | helix-turn-helix transcriptional regulator | - |
| MYG64_RS33950 (MYG64_33810) | 1287623..1288549 | - | 927 | WP_244495328 | nucleotidyltransferase and HEPN domain-containing protein | - |
| MYG64_RS33955 (MYG64_33815) | 1288958..1289926 | + | 969 | WP_112984560 | P-type conjugative transfer ATPase TrbB | virB11 |
| MYG64_RS33960 (MYG64_33820) | 1289916..1290296 | + | 381 | WP_029742474 | TrbC/VirB2 family protein | virB2 |
| MYG64_RS33965 (MYG64_33825) | 1290268..1290582 | + | 315 | WP_089045407 | conjugal transfer protein TrbD | virB3 |
| MYG64_RS33970 (MYG64_33830) | 1290593..1293043 | + | 2451 | WP_112984549 | conjugal transfer protein TrbE | virb4 |
| MYG64_RS33975 (MYG64_33835) | 1293015..1293818 | + | 804 | WP_112975498 | P-type conjugative transfer protein TrbJ | virB5 |
| MYG64_RS33980 (MYG64_33840) | 1293812..1293988 | + | 177 | WP_112975497 | entry exclusion protein TrbK | - |
| MYG64_RS33985 (MYG64_33845) | 1293993..1295177 | + | 1185 | WP_112984548 | P-type conjugative transfer protein TrbL | virB6 |
| MYG64_RS33990 (MYG64_33850) | 1295198..1295860 | + | 663 | WP_112975495 | conjugal transfer protein TrbF | virB8 |
| MYG64_RS33995 (MYG64_33855) | 1295877..1296689 | + | 813 | WP_112975494 | P-type conjugative transfer protein TrbG | virB9 |
| MYG64_RS34000 (MYG64_33860) | 1296693..1297130 | + | 438 | WP_112975493 | conjugal transfer protein TrbH | - |
| MYG64_RS34005 (MYG64_33865) | 1297143..1298438 | + | 1296 | WP_112975492 | IncP-type conjugal transfer protein TrbI | virB10 |
| MYG64_RS34010 (MYG64_33870) | 1298920..1300008 | + | 1089 | WP_112975523 | GSU2403 family nucleotidyltransferase fold protein | - |
| MYG64_RS34015 (MYG64_33875) | 1300032..1300634 | - | 603 | WP_255381712 | TraH family protein | virB1 |
| MYG64_RS34020 (MYG64_33880) | 1300648..1301814 | - | 1167 | WP_255381714 | conjugal transfer protein TraB | - |
| MYG64_RS34025 (MYG64_33885) | 1301804..1302304 | - | 501 | WP_255382351 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 6392 | GenBank | NZ_CP101519 |
| Plasmid name | NER9|pB | Incompatibility group | - |
| Plasmid size | 1586769 bp | Coordinate of oriT [Strand] | 1305751..1305794 [-] |
| Host baterium | Ensifer adhaerens strain NER9 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsC |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | AcrIIA7 |