Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105948 |
| Name | oriT_M8|pC |
| Organism | Ensifer adhaerens strain M8 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP098810 (128849..128883 [+], 35 nt) |
| oriT length | 35 nt |
| IRs (inverted repeats) | 19..24, 29..34 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 35 nt
>oriT_M8|pC
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGC
TCCAAGGGCGCAATTATACGTCGCTGATGCGACGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file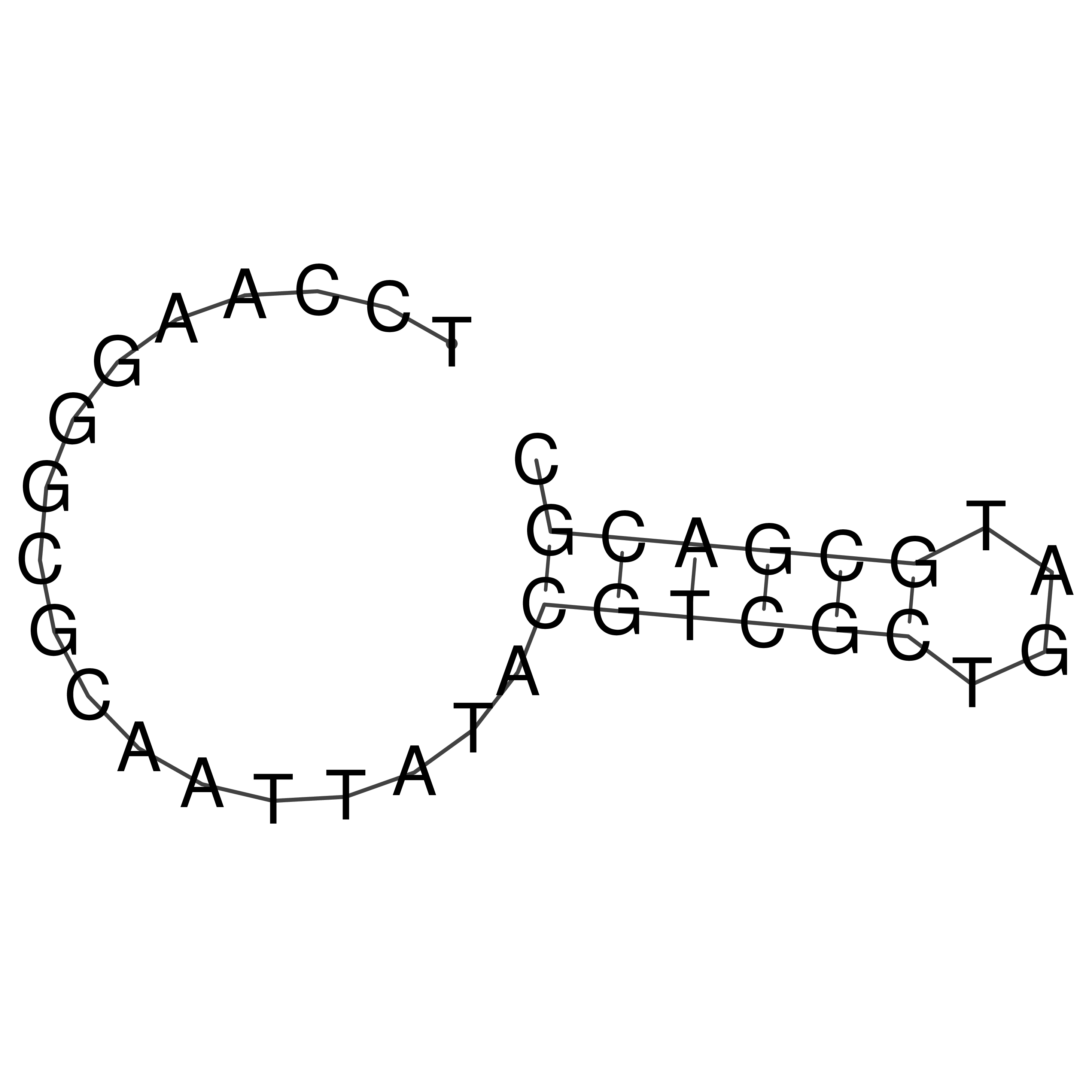
Relaxase
| ID | 4010 | GenBank | WP_252161520 |
| Name | traA_NE863_RS35540_M8|pC |
UniProt ID | _ |
| Length | 1107 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1107 a.a. Molecular weight: 123042.23 Da Isoelectric Point: 9.9099
>WP_252161520.1 Ti-type conjugative transfer relaxase TraA [Ensifer adhaerens]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKRGLLHEEFVIPAGAPEWLRSMIADR
SVSGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELTAEQNIALVRDFVELHITAKGMVADWVYHDVPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPDGNPVRNDAGKIVYELWAGSPDDFNAFRDGWFACQNRHLAMAG
LDIRVDGRSFEKQGIEVEPTIHIGVGATAIDRKAEQAKTRASAGSPKLERIELQEERRSENARRIQRRPE
IVLNLITREKSVFDERDVAKILHRYIDDAALFQSLMVRVLQSPEALRLERERIAFATGIRVPAKYTTREL
IRLEAEMANRAIWLSGRTSHGVREAVLEATFARHSRLSEEQKTAIEHVAGPERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIASRTLSSWELRWSGGRNQLDAKTIFVLDEAGMVSSRQM
AHLVETVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGKIRKA
VDAYTAEGRMIGLRLKDDAIETLIADWSRDYDPAKTTLILAHLRRDVRMLNEMARAELVERGVLGEGFAF
RTEDGHRNFAIGDQIVFLKNEGSLGVKNGMLGKVVEAAQNRIVAEIGEGEHRRQVIVEQRFYGNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLRVYYGTRSFAFAGGLIKILSRKNAKETTLDYE
KASFYRQALRFAEARGLHIVNVARTVARDRLEWTIRQKQKLADLAHRLLAIGAKLGFAKAPKPTHSQSSK
ETKPMVAGIATFSKTIEQAVADKLESDPGLKKQWEDVSSRFHLVYAQPEAAFKTVNVDAMVRDETVAKST
LAKLASQPESFGALKGKSGLLASRADKQDRERAERNVPTLAQSLGDYMRQRSQAEQRYQAEEVEVRRKVS
IDIPALSPNARQTLERVRDAIDRNDLPAALEFALADKMVKAELEGFAQAVAERFGERTFLPLAAKDTNGE
TFNAITAGMSPGQKAEVQSAWSMMRAVQKLGAHERTTEALKQANTMRQAKSQGLSLK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEYEREARTIDYTRKRGLLHEEFVIPAGAPEWLRSMIADR
SVSGASEAFWNKVEDFEKRSDAQLAKDVTIALPIELTAEQNIALVRDFVELHITAKGMVADWVYHDVPGN
PHVHLMTTLRPLTEDGFGAKKVAVTGPDGNPVRNDAGKIVYELWAGSPDDFNAFRDGWFACQNRHLAMAG
LDIRVDGRSFEKQGIEVEPTIHIGVGATAIDRKAEQAKTRASAGSPKLERIELQEERRSENARRIQRRPE
IVLNLITREKSVFDERDVAKILHRYIDDAALFQSLMVRVLQSPEALRLERERIAFATGIRVPAKYTTREL
IRLEAEMANRAIWLSGRTSHGVREAVLEATFARHSRLSEEQKTAIEHVAGPERIAAVIGRAGAGKTTMMK
AAREAWEAAGYRVVGGALAGKAAEGLEKEAGIASRTLSSWELRWSGGRNQLDAKTIFVLDEAGMVSSRQM
AHLVETVTKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQREQWMRDASLDLARGKIRKA
VDAYTAEGRMIGLRLKDDAIETLIADWSRDYDPAKTTLILAHLRRDVRMLNEMARAELVERGVLGEGFAF
RTEDGHRNFAIGDQIVFLKNEGSLGVKNGMLGKVVEAAQNRIVAEIGEGEHRRQVIVEQRFYGNLDHGYA
TTIHKSQGATVDRVKVLASLSLDRHLTYVAMTRHREDLRVYYGTRSFAFAGGLIKILSRKNAKETTLDYE
KASFYRQALRFAEARGLHIVNVARTVARDRLEWTIRQKQKLADLAHRLLAIGAKLGFAKAPKPTHSQSSK
ETKPMVAGIATFSKTIEQAVADKLESDPGLKKQWEDVSSRFHLVYAQPEAAFKTVNVDAMVRDETVAKST
LAKLASQPESFGALKGKSGLLASRADKQDRERAERNVPTLAQSLGDYMRQRSQAEQRYQAEEVEVRRKVS
IDIPALSPNARQTLERVRDAIDRNDLPAALEFALADKMVKAELEGFAQAVAERFGERTFLPLAAKDTNGE
TFNAITAGMSPGQKAEVQSAWSMMRAVQKLGAHERTTEALKQANTMRQAKSQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4017 | GenBank | WP_252161517 |
| Name | traG_NE863_RS35525_M8|pC |
UniProt ID | _ |
| Length | 660 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 660 a.a. Molecular weight: 70891.80 Da Isoelectric Point: 9.5434
>WP_252161517.1 Ti-type conjugative transfer system protein TraG [Ensifer adhaerens]
MTANKLALAGAPVVLMILVVIGMAGIEQWLSAFGKTETARLTLGRAGIAAPYVAAAATGVIFLFASAGAT
SIKFAGWGTAAGSIATILIAVVRETTRLAAFAGYVPAGKSTVSYLDPATMVGGSAALMVGCFALRIALIG
NAAFARAEPKRIRGQRALHGEADWMKMQDAAKLFADTGGIVIGERYRVDKDSTAARSFRADDAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSNEVAPMVREQRKGAARFIRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDQ
QHQTLRQVRSNLAEPEPKLRERLQEIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFSTDALADGKTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRDGEMKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPDTADYISKRCGTTT
VEIAQVSRSSQARGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHPSGKTSEARLIQPARSATSKADPGQ
MTANKLALAGAPVVLMILVVIGMAGIEQWLSAFGKTETARLTLGRAGIAAPYVAAAATGVIFLFASAGAT
SIKFAGWGTAAGSIATILIAVVRETTRLAAFAGYVPAGKSTVSYLDPATMVGGSAALMVGCFALRIALIG
NAAFARAEPKRIRGQRALHGEADWMKMQDAAKLFADTGGIVIGERYRVDKDSTAARSFRADDAETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGGLVVLDPSNEVAPMVREQRKGAARFIRILD
PKEPETGFNALDWIGRFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTALIADVCLSGHTDQ
QHQTLRQVRSNLAEPEPKLRERLQEIYDNSNSDFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPNYA
ALVSGSTFSTDALADGKTDVFINIDLKTLETHAGLARVVIGSFLNAIYNRDGEMKGRALFLLDEVARLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPDTADYISKRCGTTT
VEIAQVSRSSQARGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVG
TNRFHPSGKTSEARLIQPARSATSKADPGQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4018 | GenBank | WP_252161535 |
| Name | t4cp2_NE863_RS35620_M8|pC |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 92073.72 Da Isoelectric Point: 5.6760
>WP_252161535.1 conjugal transfer protein TrbE [Ensifer adhaerens]
MVALKRFRAAGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINTILSRLG
SGWMIQVEAVRVPTFDYPSDDRCHFPDPVTRAIDAERRAHFGREQEHFESKHALVLTYRPLESKRTALSK
YIYSDEESRKKTYADTVLFIFKNAIREVEQYFANTLSIRRMETREVAERGGERRARYDELLQFVRFCITG
ENHPIRLPDAPMYLDWIATAELEHGLTPKVENRFLGIVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIALASSQLVAYGYYTPVIVL
FDHDREGLQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYANVREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPANSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEQAQIFA
FDKGNSLLPLTLATGADHYEIGGDEHDEGRALAFCPLSDLASDADRAWATEWIEMLVALQGVAITPDHRN
AISRQVGLMADAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAERDGLSLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVSNAIPKREYYVVTPEGRRLFDMALGP
VALSFVGASGKEDLKRIRTLHSEHGRDWPIQWLQTRGVHDAVSLLDFE
MVALKRFRAAGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINTILSRLG
SGWMIQVEAVRVPTFDYPSDDRCHFPDPVTRAIDAERRAHFGREQEHFESKHALVLTYRPLESKRTALSK
YIYSDEESRKKTYADTVLFIFKNAIREVEQYFANTLSIRRMETREVAERGGERRARYDELLQFVRFCITG
ENHPIRLPDAPMYLDWIATAELEHGLTPKVENRFLGIVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVAETEDAIALASSQLVAYGYYTPVIVL
FDHDREGLQEKAEAIRRLIQAEGFGARIETLNATDAYLGSLPGNWYANVREPLINTSNLADLIPLNSVWS
GNPVAPCPFYPANSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYEQAQIFA
FDKGNSLLPLTLATGADHYEIGGDEHDEGRALAFCPLSDLASDADRAWATEWIEMLVALQGVAITPDHRN
AISRQVGLMADAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAERDGLSLGAFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLDGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCPTKICLPNGAARESGTREFYERIGFNERQIEIVSNAIPKREYYVVTPEGRRLFDMALGP
VALSFVGASGKEDLKRIRTLHSEHGRDWPIQWLQTRGVHDAVSLLDFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 134017..147517
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NE863_RS35545 (NE863_35545) | 132281..132847 | + | 567 | WP_252161521 | conjugative transfer signal peptidase TraF | - |
| NE863_RS35550 (NE863_35550) | 132837..134000 | + | 1164 | WP_252161522 | conjugal transfer protein TraB | - |
| NE863_RS35555 (NE863_35555) | 134017..134631 | + | 615 | WP_252161523 | TraH family protein | virB1 |
| NE863_RS35560 (NE863_35560) | 134940..135356 | + | 417 | WP_252161524 | DUF6088 family protein | - |
| NE863_RS35565 (NE863_35565) | 135346..136257 | + | 912 | WP_252161525 | nucleotidyl transferase AbiEii/AbiGii toxin family protein | - |
| NE863_RS35570 (NE863_35570) | 136254..136475 | - | 222 | WP_252161526 | helix-turn-helix transcriptional regulator | - |
| NE863_RS35575 (NE863_35575) | 136668..136988 | + | 321 | WP_252161527 | transcriptional repressor TraM | - |
| NE863_RS35580 (NE863_35580) | 136985..137701 | - | 717 | WP_252161600 | autoinducer binding domain-containing protein | - |
| NE863_RS35585 (NE863_35585) | 138005..139303 | - | 1299 | WP_252161528 | IncP-type conjugal transfer protein TrbI | virB10 |
| NE863_RS35590 (NE863_35590) | 139315..139752 | - | 438 | WP_252161529 | conjugal transfer protein TrbH | - |
| NE863_RS35595 (NE863_35595) | 139756..140568 | - | 813 | WP_252161530 | P-type conjugative transfer protein TrbG | virB9 |
| NE863_RS35600 (NE863_35600) | 140584..141246 | - | 663 | WP_252161531 | conjugal transfer protein TrbF | virB8 |
| NE863_RS35605 (NE863_35605) | 141268..142443 | - | 1176 | WP_252161532 | P-type conjugative transfer protein TrbL | virB6 |
| NE863_RS35610 (NE863_35610) | 142437..142637 | - | 201 | WP_252161533 | entry exclusion protein TrbK | - |
| NE863_RS35615 (NE863_35615) | 142634..143437 | - | 804 | WP_252161534 | P-type conjugative transfer protein TrbJ | virB5 |
| NE863_RS35620 (NE863_35620) | 143409..145865 | - | 2457 | WP_252161535 | conjugal transfer protein TrbE | virb4 |
| NE863_RS35625 (NE863_35625) | 145875..146174 | - | 300 | WP_252161536 | conjugal transfer protein TrbD | virB3 |
| NE863_RS35630 (NE863_35630) | 146167..146550 | - | 384 | WP_252161537 | TrbC/VirB2 family protein | virB2 |
| NE863_RS35635 (NE863_35635) | 146540..147517 | - | 978 | WP_252161538 | P-type conjugative transfer ATPase TrbB | virB11 |
| NE863_RS35640 (NE863_35640) | 147527..148150 | - | 624 | WP_252161539 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 6385 | GenBank | NZ_CP098810 |
| Plasmid name | M8|pC | Incompatibility group | - |
| Plasmid size | 148523 bp | Coordinate of oriT [Strand] | 128849..128883 [+] |
| Host baterium | Ensifer adhaerens strain M8 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | nahI |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA11b.3 |