Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105934 |
| Name | oriT_pCF10-NDM |
| Organism | Citrobacter youngae strain CF10 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP102502 (103682..103730 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..11, 18..23 (GCAAAA..TTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pCF10-NDM
AATCAGCAAAACTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCAGCAAAACTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file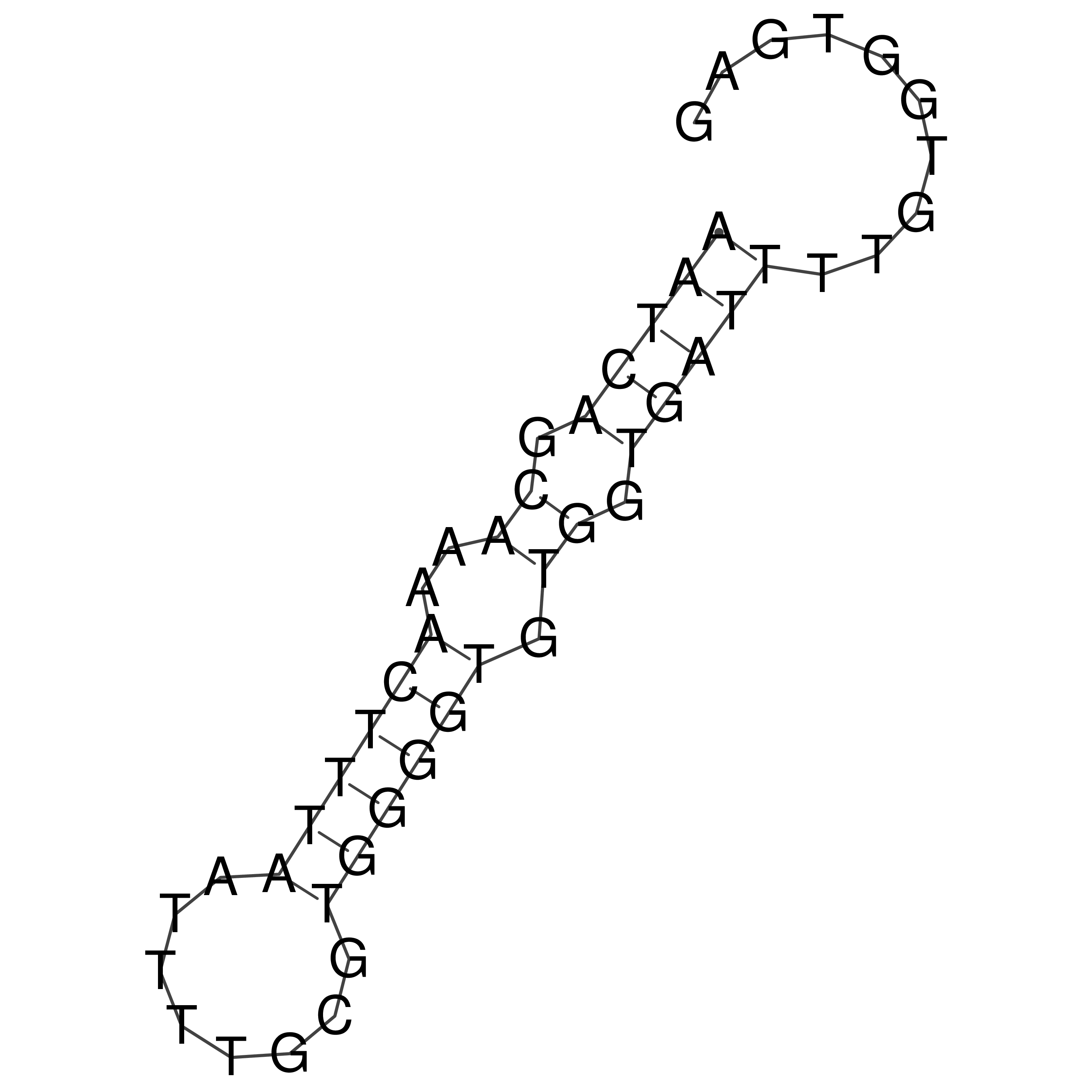
Relaxase
| ID | 4002 | GenBank | WP_057064462 |
| Name | traI_NUG39_RS25505_pCF10-NDM |
UniProt ID | A0AA40NJB4 |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 188948.08 Da Isoelectric Point: 6.0680
>WP_057064462.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKIFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQCMAEWMQT
LKATGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLASTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGAVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQVIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTPEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRGVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLAGEARPWN
PGAITGGRVWADSLPDSTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDGPAGGADVAV
RDVVRDMERIKETPASPLTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKIFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQCMAEWMQT
LKATGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLASTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTRVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGAVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVGQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQVIEKEADRQI
RSGALLSVPVSPGNGLQLLVSRQSYNAEKSILRHVLEGKEAVTPLMDKVPDAQLAGLTDGQQNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERPDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTPEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVTLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRGVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKFISPGRKYPQPHVALPAFDRNGRQAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLAGEARPWN
PGAITGGRVWADSLPDSTGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDGPAGGADVAV
RDVVRDMERIKETPASPLTLPDAPEERRRDEAVSQVVRENVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 4007 | GenBank | WP_057064463 |
| Name | traD_NUG39_RS25510_pCF10-NDM |
UniProt ID | _ |
| Length | 722 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 722 a.a. Molecular weight: 80873.32 Da Isoelectric Point: 5.1609
>WP_057064463.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGVIVALTLSIKLTWQTFVNGMVWYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEASAENTGQPQQPQQLTP
APRAADAPSASASAAGAAGTGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQAEQDELSPQERQR
REEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGVIVALTLSIKLTWQTFVNGMVWYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVIAPEASAENTGQPQQPQQLTP
APRAADAPSASASAAGAAGTGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAWQAEQDELSPQERQR
REEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 4008 | GenBank | WP_008324134 |
| Name | traC_NUG39_RS25585_pCF10-NDM |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98841.51 Da Isoelectric Point: 5.7676
>WP_008324134.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacterales]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 78425..104288
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NUG39_RS25510 (NUG39_25510) | 78425..80593 | - | 2169 | WP_057064463 | type IV conjugative transfer system coupling protein TraD | virb4 |
| NUG39_RS25515 (NUG39_25515) | 80712..81512 | - | 801 | WP_226999298 | hypothetical protein | - |
| NUG39_RS25520 (NUG39_25520) | 81549..82025 | - | 477 | WP_071886900 | hypothetical protein | - |
| NUG39_RS25525 (NUG39_25525) | 82113..83081 | - | 969 | WP_000654812 | IS5-like element IS903B family transposase | - |
| NUG39_RS25530 (NUG39_25530) | 83099..85927 | - | 2829 | WP_257749357 | conjugal transfer mating-pair stabilization protein TraG | traG |
| NUG39_RS25535 (NUG39_25535) | 85927..87303 | - | 1377 | WP_086551404 | conjugal transfer pilus assembly protein TraH | traH |
| NUG39_RS25540 (NUG39_25540) | 87300..87845 | - | 546 | WP_045379409 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| NUG39_RS25545 (NUG39_25545) | 87835..88083 | - | 249 | WP_045379412 | type-F conjugative transfer system pilin chaperone TraQ | - |
| NUG39_RS25550 (NUG39_25550) | 88100..88849 | - | 750 | WP_008324127 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| NUG39_RS25555 (NUG39_25555) | 88880..89068 | - | 189 | WP_008324128 | conjugal transfer protein TrbE | - |
| NUG39_RS25560 (NUG39_25560) | 89100..90953 | - | 1854 | WP_016479983 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| NUG39_RS25565 (NUG39_25565) | 90950..91579 | - | 630 | WP_008324130 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| NUG39_RS25570 (NUG39_25570) | 91596..92582 | - | 987 | WP_032160671 | conjugal transfer pilus assembly protein TraU | traU |
| NUG39_RS25575 (NUG39_25575) | 92585..93232 | - | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| NUG39_RS25580 (NUG39_25580) | 93232..93579 | - | 348 | WP_008324133 | type-F conjugative transfer system protein TrbI | - |
| NUG39_RS25585 (NUG39_25585) | 93576..96206 | - | 2631 | WP_008324134 | type IV secretion system protein TraC | virb4 |
| NUG39_RS25590 (NUG39_25590) | 96199..96765 | - | 567 | WP_008324135 | conjugal transfer pilus-stabilizing protein TraP | - |
| NUG39_RS25595 (NUG39_25595) | 96755..97156 | - | 402 | WP_257749361 | hypothetical protein | - |
| NUG39_RS25600 (NUG39_25600) | 97140..97325 | - | 186 | WP_008324139 | hypothetical protein | - |
| NUG39_RS25605 (NUG39_25605) | 97329..97553 | - | 225 | WP_016247532 | TraR/DksA C4-type zinc finger protein | - |
| NUG39_RS25610 (NUG39_25610) | 97673..98212 | - | 540 | WP_016247533 | type IV conjugative transfer system lipoprotein TraV | traV |
| NUG39_RS25615 (NUG39_25615) | 98234..99628 | - | 1395 | WP_008324156 | F-type conjugal transfer pilus assembly protein TraB | traB |
| NUG39_RS25620 (NUG39_25620) | 99615..100358 | - | 744 | WP_008324157 | type-F conjugative transfer system secretin TraK | traK |
| NUG39_RS25625 (NUG39_25625) | 100345..100911 | - | 567 | WP_016247535 | type IV conjugative transfer system protein TraE | traE |
| NUG39_RS25630 (NUG39_25630) | 100926..101231 | - | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| NUG39_RS25635 (NUG39_25635) | 101382..101732 | - | 351 | WP_016247536 | type IV conjugative transfer system pilin TraA | - |
| NUG39_RS25640 (NUG39_25640) | 101790..102014 | - | 225 | WP_008324165 | TraY domain-containing protein | - |
| NUG39_RS25645 (NUG39_25645) | 102101..102790 | - | 690 | WP_008324166 | hypothetical protein | - |
| NUG39_RS25650 (NUG39_25650) | 102979..103371 | - | 393 | WP_008324167 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| NUG39_RS25655 (NUG39_25655) | 103803..104288 | + | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| NUG39_RS25660 (NUG39_25660) | 104321..104650 | - | 330 | WP_011977736 | DUF5983 family protein | - |
| NUG39_RS25665 (NUG39_25665) | 104683..105504 | - | 822 | WP_032454971 | DUF932 domain-containing protein | - |
| NUG39_RS25670 (NUG39_25670) | 106320..106719 | - | 400 | Protein_115 | hypothetical protein | - |
Host bacterium
| ID | 6371 | GenBank | NZ_CP102502 |
| Plasmid name | pCF10-NDM | Incompatibility group | IncFIB |
| Plasmid size | 138196 bp | Coordinate of oriT [Strand] | 103682..103730 [+] |
| Host baterium | Citrobacter youngae strain CF10 |
Cargo genes
| Drug resistance gene | blaNDM-1 |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |