Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105819 |
| Name | oriT_pRvVAR031b |
| Organism | Agrobacterium vitis strain VAR03-1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP023271 (40164..40207 [-], 44 nt) |
| oriT length | 44 nt |
| IRs (inverted repeats) | 21..27, 32..38 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 44 nt
>oriT_pRvVAR031b
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
GGATCCAAGGGCGCAATTATACGTCGCTGATGCGACGTGCTTGC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file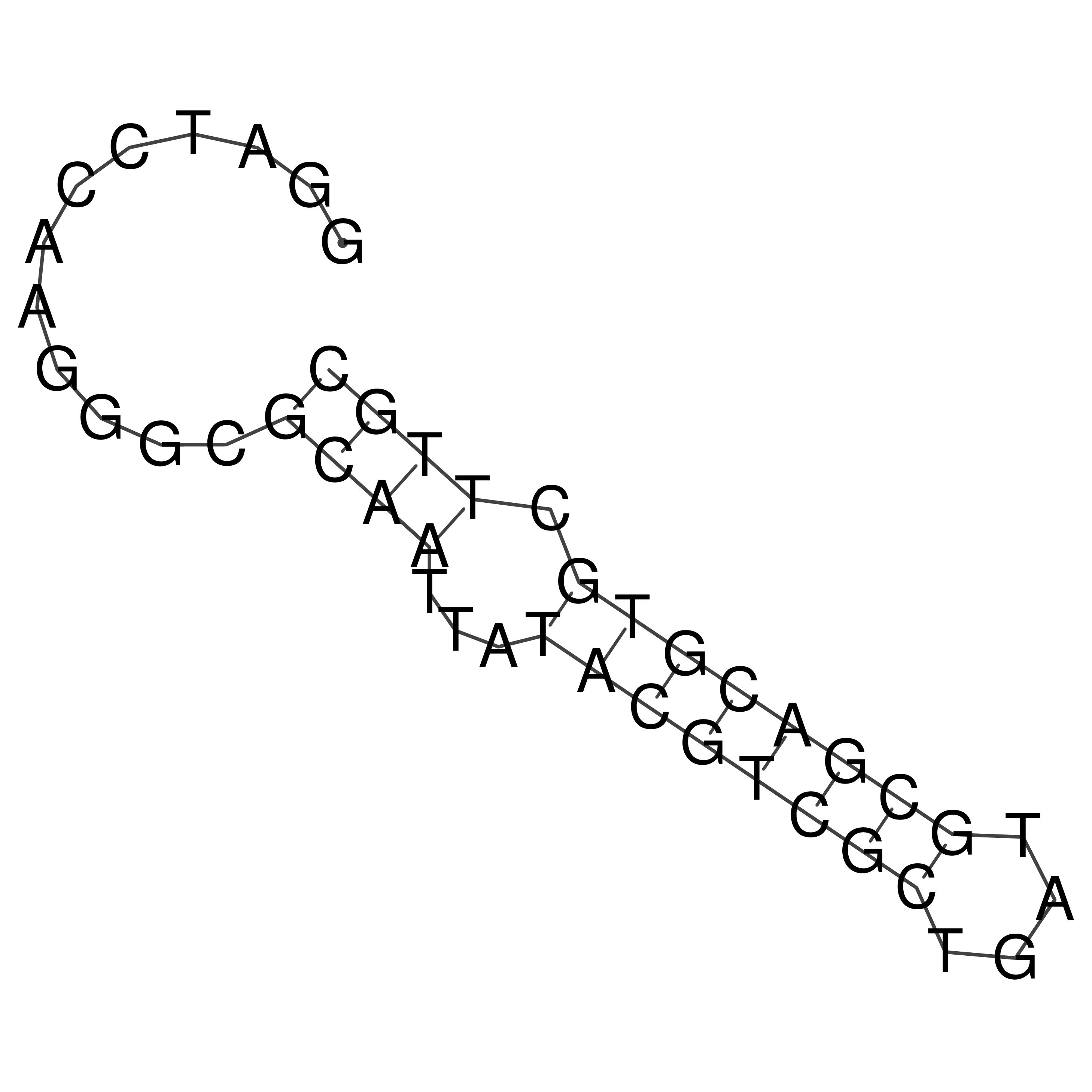
Relaxase
| ID | 3929 | GenBank | WP_180642278 |
| Name | traA_RvVAT039_RS24250_pRvVAR031b |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123485.17 Da Isoelectric Point: 10.0423
>WP_180642278.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium vitis]
MAIAHFSASIVSRGGGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRALIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIHAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEEGFGSKKVAVTAKGGQPVRTKSGKILYELWAGSTGDFNVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGIDLEPTIHLGVGAKAIERKAQQQGVRPELERIELNEQRRSENARRILNNPAIVLDV
ITREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKLPARYSTRAMIRLEA
TMVRQTLWLSNRDGHSVSEAALDATFRRHERLSQEQKAAIERITGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDHKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLIGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREGWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNNDYDQSKTTLILAHLRRDVRTLNIRAREKLVERGIVGEGHVFKTADG
VRQFDAGDQIVFLKNEGLLGVKNGMIAHVIEAAPSRIVAVIGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGHQSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAESRGLNIVQVARTLVRDRLDWTLRQKTRLVDLGQRLAVFAERLGLHHPLKTQTKKEAAPMVAG
IKTFPGSVADTVENKLDTDPALRQQWEEVSTRFAYVFADPETAFRAMNFDTVPADKEKAKLALQTLTTAP
ASIGPLKGKTGILASKTEREDRRVADLNVPALKRDLEQYLRMRETAVQRLQADEQVLRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERTMLTNAAREPSGKLFDKLSDG
MQPEQKERLKEAWPIMRTSQQLAAHERTVQTLRQAEDLRLAQRQTPVMKQ
MAIAHFSASIVSRGGGRSAVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFVLPADAPKWVRALIADR
SVSGAVEAFWNKVEAFEKRSDAQLARDLTIALPLELTPEQNIALVRDFVEKHIHAKGMVADWVYHDNPGN
PHIHLMTTLRPLTEEGFGSKKVAVTAKGGQPVRTKSGKILYELWAGSTGDFNVLRDGWFERLNHHLALGG
IDLKIDGRSYEKQGIDLEPTIHLGVGAKAIERKAQQQGVRPELERIELNEQRRSENARRILNNPAIVLDV
ITREKSVFDERDVAKVLHRYVDDPAVFQQLMLRIILNPEVLRLQRDTIEFATGEKLPARYSTRAMIRLEA
TMVRQTLWLSNRDGHSVSEAALDATFRRHERLSQEQKAAIERITGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDHKTVFVMDEAGMVASKQMAGFVD
AVVRAGAKIVLIGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREGWMRKASLDLARGNVEKALTAYD
ANVRITGTRLKAEAVERLIGDWNNDYDQSKTTLILAHLRRDVRTLNIRAREKLVERGIVGEGHVFKTADG
VRQFDAGDQIVFLKNEGLLGVKNGMIAHVIEAAPSRIVAVIGEGDQRRQVIVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGHQSFAFNGGLAKVLSRKNAKETTLDYERGKLY
REALRFAESRGLNIVQVARTLVRDRLDWTLRQKTRLVDLGQRLAVFAERLGLHHPLKTQTKKEAAPMVAG
IKTFPGSVADTVENKLDTDPALRQQWEEVSTRFAYVFADPETAFRAMNFDTVPADKEKAKLALQTLTTAP
ASIGPLKGKTGILASKTEREDRRVADLNVPALKRDLEQYLRMRETAVQRLQADEQVLRQRVSIDIPALSP
AAHVVLERVRDAIDRNDLPAAMAYALSNRETKLEIDGFNQAVTERFGERTMLTNAAREPSGKLFDKLSDG
MQPEQKERLKEAWPIMRTSQQLAAHERTVQTLRQAEDLRLAQRQTPVMKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3945 | GenBank | WP_180642233 |
| Name | t4cp2_RvVAT039_RS24095_pRvVAR031b |
UniProt ID | _ |
| Length | 822 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 822 a.a. Molecular weight: 91702.50 Da Isoelectric Point: 5.4869
>WP_180642233.1 conjugal transfer protein TrbE [Agrobacterium vitis]
MVALKSLRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNDVSRQINAILSRLG
SGWMIQVEAVRVPTGDYPTEAESHFPDPVTRAIDDERRTHFQTERGHYESRHALILTWRPPEPRRSGLTR
YVYSDTTSRSATYADTMLQSFLTSIREVEQYLANVVSIRRMMTRETPERGGFRVARYDELFQFIRFCVTG
ENHPVRLPDISMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDPTRLQEKCEAIRKLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFFPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTLGLNGDHYQIGGDIGPSAGGEVKALAFCPLAELSTDGDRAWASEWIEMLVALQGVTISP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEIEDL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNM
ALGPVALSFVGATGKEDLKQIRSLHSEHGAAWPCPWLQQRGIAHAETLFSAE
MVALKSLRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNDVSRQINAILSRLG
SGWMIQVEAVRVPTGDYPTEAESHFPDPVTRAIDDERRTHFQTERGHYESRHALILTWRPPEPRRSGLTR
YVYSDTTSRSATYADTMLQSFLTSIREVEQYLANVVSIRRMMTRETPERGGFRVARYDELFQFIRFCVTG
ENHPVRLPDISMYLDWLVTAELQHGLTPMVENRFLGVVAIDGLPAESWPGILNTLDRMPLTYRWSSRFVF
LDAEEARARLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVI
FEEDPTRLQEKCEAIRKLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLIPLNSVWS
GSPTAPCPFFPPASPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYADAQVFA
FDKGGSMLPLTLGLNGDHYQIGGDIGPSAGGEVKALAFCPLAELSTDGDRAWASEWIEMLVALQGVTISP
DYRNAISRQLALMAESRGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLALGAFQCFEIEDL
MNMGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSIS
DAERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKREYYVVSPEGRRLFNM
ALGPVALSFVGATGKEDLKQIRSLHSEHGAAWPCPWLQQRGIAHAETLFSAE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3946 | GenBank | WP_071207755 |
| Name | traG_RvVAT039_RS24265_pRvVAR031b |
UniProt ID | _ |
| Length | 656 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 656 a.a. Molecular weight: 70396.46 Da Isoelectric Point: 9.3253
>WP_071207755.1 Ti-type conjugative transfer system protein TraG [Agrobacterium vitis]
MTVGKLLLAVIPATAMIVVVVFMPGIEHWLAAFGKTAQTKLMLGRIGLALPYATAAAIGTICLFAANGAA
GIKRAGWGVVGGNGVAILIALLREGVRLAGIAGDIPEGQSVLGYVDPPTTLGASTTFLAGVFALRVAMNG
NAAFAKAAPRRIGGKRAVHGEADWMKVQEAAKLFPNSGGIVVGERYRVGRDSVAAVSFRADDPSTWGAGG
KNPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSSEVAPMVSEHRRKAGRKVIILD
PSASGVGFNALDWIGRHGSTKEEDIVSVATWIMTDNPRSASARDDFFRASAMQLLTALIADVCLSGHTDE
EDQTLRRVRKNLSEPEPQLRARLTKIYEQSESDFVKENVSVFVNMTPETFCGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGDVKGRALFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSRQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRRKDMSASVG
ENRFHKPITEGVKSYKAAPTTDTEAT
MTVGKLLLAVIPATAMIVVVVFMPGIEHWLAAFGKTAQTKLMLGRIGLALPYATAAAIGTICLFAANGAA
GIKRAGWGVVGGNGVAILIALLREGVRLAGIAGDIPEGQSVLGYVDPPTTLGASTTFLAGVFALRVAMNG
NAAFAKAAPRRIGGKRAVHGEADWMKVQEAAKLFPNSGGIVVGERYRVGRDSVAAVSFRADDPSTWGAGG
KNPLLCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGGGLVVLDPSSEVAPMVSEHRRKAGRKVIILD
PSASGVGFNALDWIGRHGSTKEEDIVSVATWIMTDNPRSASARDDFFRASAMQLLTALIADVCLSGHTDE
EDQTLRRVRKNLSEPEPQLRARLTKIYEQSESDFVKENVSVFVNMTPETFCGVYANAVKETHWLSYPNYA
ALVSGDSFSTDDLADGGTDIFIALDLKVLEAHPGLARVVIGSLLNAIYNRNGDVKGRALFLLDEVARLGY
LRILETARDAGRKYGITLTMIFQSIGQMREAYGGRDATSKWFESASWISFAAINDPDTADYISKRCGDTT
VEVDQTNRSSGMKGSSRSRSRQLSRRPLILPHEVLRMRADEQIVFTSGNPPLRCGRAIWFRRKDMSASVG
ENRFHKPITEGVKSYKAAPTTDTEAT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4264..13956
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| RvVAT039_RS24065 (RvVAR031_pl05490) | 984..1979 | - | 996 | WP_180642225 | plasmid partitioning protein RepB | - |
| RvVAT039_RS24070 (RvVAR031_pl05500) | 2064..3254 | - | 1191 | WP_180642227 | plasmid partitioning protein RepA | - |
| RvVAT039_RS24075 (RvVAR031_pl05510) | 3632..4267 | + | 636 | WP_180642228 | acyl-homoserine-lactone synthase | - |
| RvVAT039_RS24080 (RvVAR031_pl05520) | 4264..5235 | + | 972 | WP_180642230 | P-type conjugative transfer ATPase TrbB | virB11 |
| RvVAT039_RS24085 (RvVAR031_pl05530) | 5225..5629 | + | 405 | WP_180642232 | conjugal transfer pilin TrbC | virB2 |
| RvVAT039_RS24090 (RvVAR031_pl05540) | 5622..5921 | + | 300 | WP_020046779 | conjugal transfer protein TrbD | virB3 |
| RvVAT039_RS24095 (RvVAR031_pl05550) | 5931..8399 | + | 2469 | WP_180642233 | conjugal transfer protein TrbE | virb4 |
| RvVAT039_RS24100 (RvVAR031_pl05560) | 8371..9180 | + | 810 | WP_180642235 | P-type conjugative transfer protein TrbJ | virB5 |
| RvVAT039_RS24105 (RvVAR031_pl05570) | 9177..9404 | + | 228 | WP_180642237 | entry exclusion protein TrbK | - |
| RvVAT039_RS24110 (RvVAR031_pl05580) | 9398..10585 | + | 1188 | WP_180642239 | P-type conjugative transfer protein TrbL | virB6 |
| RvVAT039_RS24115 (RvVAR031_pl05590) | 10603..11280 | + | 678 | WP_180642241 | conjugal transfer protein TrbF | virB8 |
| RvVAT039_RS24120 (RvVAR031_pl05600) | 11355..12149 | + | 795 | WP_234616830 | P-type conjugative transfer protein TrbG | virB9 |
| RvVAT039_RS24125 (RvVAR031_pl05610) | 12149..12625 | + | 477 | WP_180642245 | conjugal transfer protein TrbH | - |
| RvVAT039_RS24130 (RvVAR031_pl05620) | 12637..13956 | + | 1320 | WP_180642248 | IncP-type conjugal transfer protein TrbI | virB10 |
| RvVAT039_RS24135 (RvVAR031_pl05630) | 14093..14995 | - | 903 | WP_156551308 | LysR family transcriptional regulator | - |
| RvVAT039_RS24140 (RvVAR031_pl05640) | 15248..16021 | + | 774 | WP_156551382 | nopaline ABC transporter ATP-binding protein NocP | - |
| RvVAT039_RS24145 (RvVAR031_pl05650) | 16087..16938 | + | 852 | WP_156551309 | nopaline ABC transporter substrate-binding protein NocT | - |
| RvVAT039_RS24150 (RvVAR031_pl05660) | 17011..17721 | + | 711 | WP_045025159 | nopaline ABC transporter permease NocQ | - |
| RvVAT039_RS24155 (RvVAR031_pl05670) | 17723..18448 | + | 726 | WP_080868320 | nopaline ABC transporter permease NocM | - |
Host bacterium
| ID | 6256 | GenBank | NZ_AP023271 |
| Plasmid name | pRvVAR031b | Incompatibility group | - |
| Plasmid size | 161351 bp | Coordinate of oriT [Strand] | 40164..40207 [-] |
| Host baterium | Agrobacterium vitis strain VAR03-1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |