Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105704 |
| Name | oriT_pEcl3-2 |
| Organism | Enterobacter hormaechei strain Eho-3 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP047751 (55910..55996 [-], 87 nt) |
| oriT length | 87 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 87 nt
>oriT_pEcl3-2
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTGATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
GGGTGTCGGGGCGGAGCCCTGACCAGGTGGCATTTGTGATATCGTGCGTGCGCGGTATTACAAATGCACATCCTGTCCCGTCTTTCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file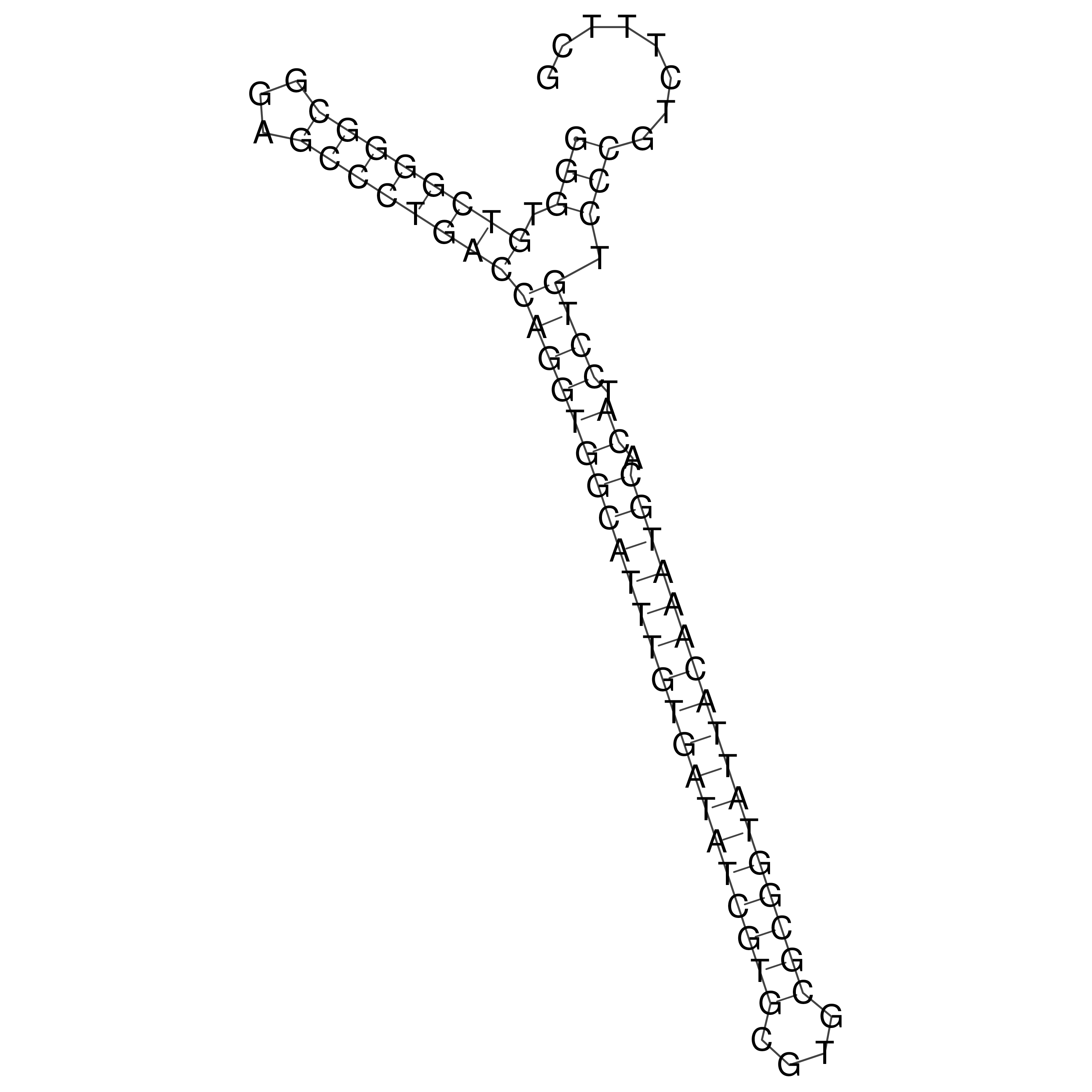
Relaxase
| ID | 3862 | GenBank | WP_022650008 |
| Name | Relaxase_GVI64_RS25745_pEcl3-2 |
UniProt ID | A0A4Q2QTK9 |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 102314.37 Da Isoelectric Point: 8.2756
>WP_022650008.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacterales]
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVRASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKTKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWKRGRNQSLREYIADTAIAGLRESPVTDWSSLHQRFAKEGLFLTKE
SGELKVKDGWDSERPGVTLSAFGHAFDTDKLIRKLGEFVPPVQDIFSQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYAIARLRAPLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDSTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFQQVPPAEQFTGGSLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIESLIGSGEFTWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGSFVSVPADIFERVKPASRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQAIHDECRIRKARIRIEHRDPLVRKLHYHIAELQRMQALIALKETMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDNRCVVICEPGGTPMYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNSGDYDALKRDMVKVAPVLFSRSPSVAMAPEGNNEQFNTAFAKM
VAWHNVNHRDEGEYQISRPEVDQVRVESEMRFAEMTNQGSNTEAVRNESGPNWKPPSPL
MIPVIPPKRQDGKSSFGDLVSYVSVRDEPRDDDLMEAVRASGTKPEMPHSSRFSRLVDYATKLRDESFIS
LVDVMPDGGEWVNFYGVTCFHNCTSIESAAEEMEFSARKTKFAHNKSDPVFHYILSWQSHESPRPEQIYD
SVSHTLSRLGLAGHQFVAAVHTDTDNLHVHIAVNRVHPETGYLNRLAYSQEKLSKACRELELKHGFSPDN
GCYVIAPDNRIVRRTSVERDRQSAWKRGRNQSLREYIADTAIAGLRESPVTDWSSLHQRFAKEGLFLTKE
SGELKVKDGWDSERPGVTLSAFGHAFDTDKLIRKLGEFVPPVQDIFSQVPAVGRYEPDKIIMPSRPERMT
EKESLSDYAIARLRAPLIALDQDPTQRTIQSVHTLLAQSGLYLKEQHDRLVICDGYDSTRTPVRAERVWP
ALTKAILDSYKGGWQPVPKDIFQQVPPAEQFTGGSLEATPVSDREWRKLRTGSGPQGALKREIFSDKESL
WGYAVSHCRQDIESLIGSGEFTWERCHELLARQGLLLMREQQGLVVMDAYNHEQTPVKASHVHPDLTLAR
AEPHAGSFVSVPADIFERVKPASRYNPELAVSDRDIPGMKRDPELRRQRREARAAAREDLKARYAAWRAN
WQRPDLHGRERYQAIHDECRIRKARIRIEHRDPLVRKLHYHIAELQRMQALIALKETMKAERAELIGQGK
WYPPSYRQWVEAEAVKGDKAAISQLRGWDYRDRRGNAVKSTTDNRCVVICEPGGTPMYSGVPGLRASLKK
NGRVQFRDPETGRHICTDYGDRVIFRNSGDYDALKRDMVKVAPVLFSRSPSVAMAPEGNNEQFNTAFAKM
VAWHNVNHRDEGEYQISRPEVDQVRVESEMRFAEMTNQGSNTEAVRNESGPNWKPPSPL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A4Q2QTK9 |
T4CP
| ID | 3875 | GenBank | WP_022650006 |
| Name | trbC_GVI64_RS25760_pEcl3-2 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87660.94 Da Isoelectric Point: 6.8660
>WP_022650006.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacterales]
MNYTPVNQALMKRNAWNSPVLELLRDHLLYAGIMAGSVVAGFIWPLAIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSLFRRFPSLFQYETTQESKGKGIFYLGYRRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IHNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAELYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHRLE
GLDSEALEITKYKGPFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIVMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTITNAAGQEARANMVSLQRRDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKAAKAEPWKIIDTFQQRLANRQEAHNLMTEYDTDHRGREENLWEEALEVIRTTTMAERTI
RYITLNKPESEPTQVSAEEIQLAPDAILRTLTLPQPAYIPPSRYRNEPFISPKLPDSPHPDMEWPE
MNYTPVNQALMKRNAWNSPVLELLRDHLLYAGIMAGSVVAGFIWPLAIPLLLLLAIVASISFGTHRWRMP
MRMPVHLNKVDPSEDRKVRRSLFRRFPSLFQYETTQESKGKGIFYLGYRRMNDIGRELWLTIDDLTRHVM
FFASTGGGKTETIYAWMLNSFCWGRGFTFVDGKAQNDTARTCYYLARRFGREDDVEYINFMNSGMSRSEM
IHNGDKSRPQSHQWNPFCYSTEAFVAETMQSMLPTNVQGGEWQSRAIAMNKALVFGTKFYCVRENKTMSL
QLLREFMPLEKLAELYCRAIDDQWPEEAVSPLYNYLVDVPGFDMATVRQPSAWTEEPRKQHSYLTGQFLE
TFNTFTETFGNIFAEDAGDIDIRDSIHSDRILLVLIPAMNTSQHTTSALGRMFVTQQSMILARDLGHRLE
GLDSEALEITKYKGPFPYMNYLDEVGAYYTERIAVQATQVRSLDFSIVMMSQDQERIESQTSATNVATLM
QNAGTKVAGKIVSDDKTAKTITNAAGQEARANMVSLQRRDGIIGTSWIDGDHINIQMENKINVQDLIKLQ
PGENYTVFQGDPVPGASFYIEPQDKTCNAPVIINRYITINPPNLKQLRKLVPRTAQRRLPAPENVSKIIG
VLTEKPSRRRKAAKAEPWKIIDTFQQRLANRQEAHNLMTEYDTDHRGREENLWEEALEVIRTTTMAERTI
RYITLNKPESEPTQVSAEEIQLAPDAILRTLTLPQPAYIPPSRYRNEPFISPKLPDSPHPDMEWPE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 62119..99025
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GVI64_RS25750 (GVI64_25695) | 59135..59338 | - | 204 | WP_123839396 | hypothetical protein | - |
| GVI64_RS25755 (GVI64_25700) | 59472..59741 | - | 270 | WP_022650007 | hypothetical protein | - |
| GVI64_RS25760 (GVI64_25705) | 59815..62115 | - | 2301 | WP_022650006 | F-type conjugative transfer protein TrbC | - |
| GVI64_RS25765 (GVI64_25710) | 62119..63375 | - | 1257 | WP_126530067 | hypothetical protein | trbB |
| GVI64_RS25770 (GVI64_25715) | 63356..64564 | - | 1209 | WP_022650004 | hypothetical protein | trbA |
| GVI64_RS25775 (GVI64_25720) | 64885..65517 | - | 633 | WP_022650003 | plasmid IncI1-type surface exclusion protein ExcA | - |
| GVI64_RS25780 (GVI64_25725) | 65579..67738 | - | 2160 | WP_022650002 | DotA/TraY family protein | traY |
| GVI64_RS25785 (GVI64_25730) | 67796..68338 | - | 543 | WP_022650001 | conjugal transfer protein TraX | - |
| GVI64_RS25790 (GVI64_25735) | 68335..69537 | - | 1203 | WP_022650000 | conjugal transfer protein TraW | traW |
| GVI64_RS25795 (GVI64_25740) | 69550..70122 | - | 573 | WP_154590113 | conjugal transfer protein TraV | traV |
| GVI64_RS25800 (GVI64_25745) | 70119..73172 | - | 3054 | WP_022649998 | hypothetical protein | traU |
| GVI64_RS25805 (GVI64_25750) | 73259..73984 | - | 726 | WP_063864836 | hypothetical protein | traT |
| GVI64_RS25810 (GVI64_25755) | 74168..74572 | - | 405 | WP_022649996 | DUF6750 family protein | traR |
| GVI64_RS25815 (GVI64_25760) | 74607..75131 | - | 525 | WP_022649995 | conjugal transfer protein TraQ | traQ |
| GVI64_RS25820 (GVI64_25765) | 75131..75904 | - | 774 | WP_123839398 | conjugal transfer protein TraP | traP |
| GVI64_RS25825 (GVI64_25770) | 75885..77192 | - | 1308 | WP_022649993 | conjugal transfer protein TraO | traO |
| GVI64_RS25830 (GVI64_25775) | 77192..78172 | - | 981 | WP_022649992 | DotH/IcmK family type IV secretion protein | traN |
| GVI64_RS25835 (GVI64_25780) | 78183..78800 | - | 618 | WP_179297403 | DotI/IcmL family type IV secretion protein | traM |
| GVI64_RS25840 (GVI64_25785) | 78890..79240 | - | 351 | WP_022649990 | hypothetical protein | traL |
| GVI64_RS25845 (GVI64_25790) | 79258..82800 | - | 3543 | WP_032635082 | LPD7 domain-containing protein | - |
| GVI64_RS25850 (GVI64_25795) | 82872..83159 | - | 288 | WP_022649988 | hypothetical protein | traK |
| GVI64_RS25855 (GVI64_25800) | 83156..84316 | - | 1161 | WP_022649987 | plasmid transfer ATPase TraJ | virB11 |
| GVI64_RS25860 (GVI64_25805) | 84329..85135 | - | 807 | WP_022649986 | type IV secretory system conjugative DNA transfer family protein | traI |
| GVI64_RS25865 (GVI64_25810) | 85132..85605 | - | 474 | WP_022649985 | DotD/TraH family lipoprotein | - |
| GVI64_RS25870 (GVI64_25815) | 85676..86092 | - | 417 | WP_123839394 | hypothetical protein | - |
| GVI64_RS25875 (GVI64_25820) | 86201..87406 | - | 1206 | WP_022649984 | conjugal transfer protein TraF | - |
| GVI64_RS25880 (GVI64_25825) | 87512..88348 | - | 837 | WP_022649983 | hypothetical protein | traE |
| GVI64_RS25885 (GVI64_25830) | 88495..88971 | - | 477 | WP_022649982 | hypothetical protein | - |
| GVI64_RS25890 (GVI64_25835) | 88991..90151 | - | 1161 | WP_022649981 | site-specific integrase | - |
| GVI64_RS25895 (GVI64_25840) | 90191..91729 | - | 1539 | WP_197377751 | IS66-like element ISKpn24 family transposase | - |
| GVI64_RS25900 (GVI64_25845) | 91778..92125 | - | 348 | WP_000612626 | IS66 family insertion sequence element accessory protein TnpB | - |
| GVI64_RS25905 (GVI64_25850) | 92122..92526 | - | 405 | WP_003031976 | transposase | - |
| GVI64_RS25910 (GVI64_25855) | 93194..94528 | - | 1335 | WP_045346679 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| GVI64_RS25915 (GVI64_25860) | 94525..95199 | - | 675 | WP_022649979 | prepilin peptidase | - |
| GVI64_RS25920 (GVI64_25865) | 95184..95666 | - | 483 | WP_045261878 | lytic transglycosylase domain-containing protein | virB1 |
| GVI64_RS25925 (GVI64_25870) | 95751..96365 | - | 615 | WP_022649977 | type 4 pilus major pilin | - |
| GVI64_RS25930 (GVI64_25875) | 96379..97473 | - | 1095 | WP_032623575 | type II secretion system F family protein | - |
| GVI64_RS25935 (GVI64_25880) | 97466..99025 | - | 1560 | WP_022649975 | ATPase, T2SS/T4P/T4SS family | virB11 |
| GVI64_RS25940 (GVI64_25885) | 99028..99543 | - | 516 | WP_022649974 | type IV pilus biogenesis protein PilP | - |
| GVI64_RS25945 (GVI64_25890) | 99527..100831 | - | 1305 | WP_022649973 | type 4b pilus protein PilO2 | - |
| GVI64_RS25950 (GVI64_25895) | 100833..102509 | - | 1677 | WP_022649972 | PilN family type IVB pilus formation outer membrane protein | - |
| GVI64_RS25955 (GVI64_25900) | 102524..102961 | - | 438 | WP_022649971 | type IV pilus biogenesis protein PilM | - |
Host bacterium
| ID | 6141 | GenBank | NZ_CP047751 |
| Plasmid name | pEcl3-2 | Incompatibility group | - |
| Plasmid size | 109160 bp | Coordinate of oriT [Strand] | 55910..55996 [-] |
| Host baterium | Enterobacter hormaechei strain Eho-3 |
Cargo genes
| Drug resistance gene | blaLAP-2, qnrS1 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIC1, AcrIIA7 |