Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105702 |
| Name | oriT_pEclE3-3 |
| Organism | Enterobacter hormaechei strain Eho-E3 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP049024 (42252..42352 [+], 101 nt) |
| oriT length | 101 nt |
| IRs (inverted repeats) | 80..85, 91..96 (AAAAAA..TTTTTT) 20..26, 38..44 (TAAATCA..TGATTTA) |
| Location of nic site | 62..63 |
| Conserved sequence flanking the nic site |
GGTGTATAGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 101 nt
>oriT_pEclE3-3
TATTTATTTTTTTATCTTTTAAATCAGTATGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
TATTTATTTTTTTATCTTTTAAATCAGTATGATAGCGTGATTTATCGCGCTGCGTTAGGTGTATAGCAGGTTAAGGGATAAAAAATCATCTTTTTTGGTAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file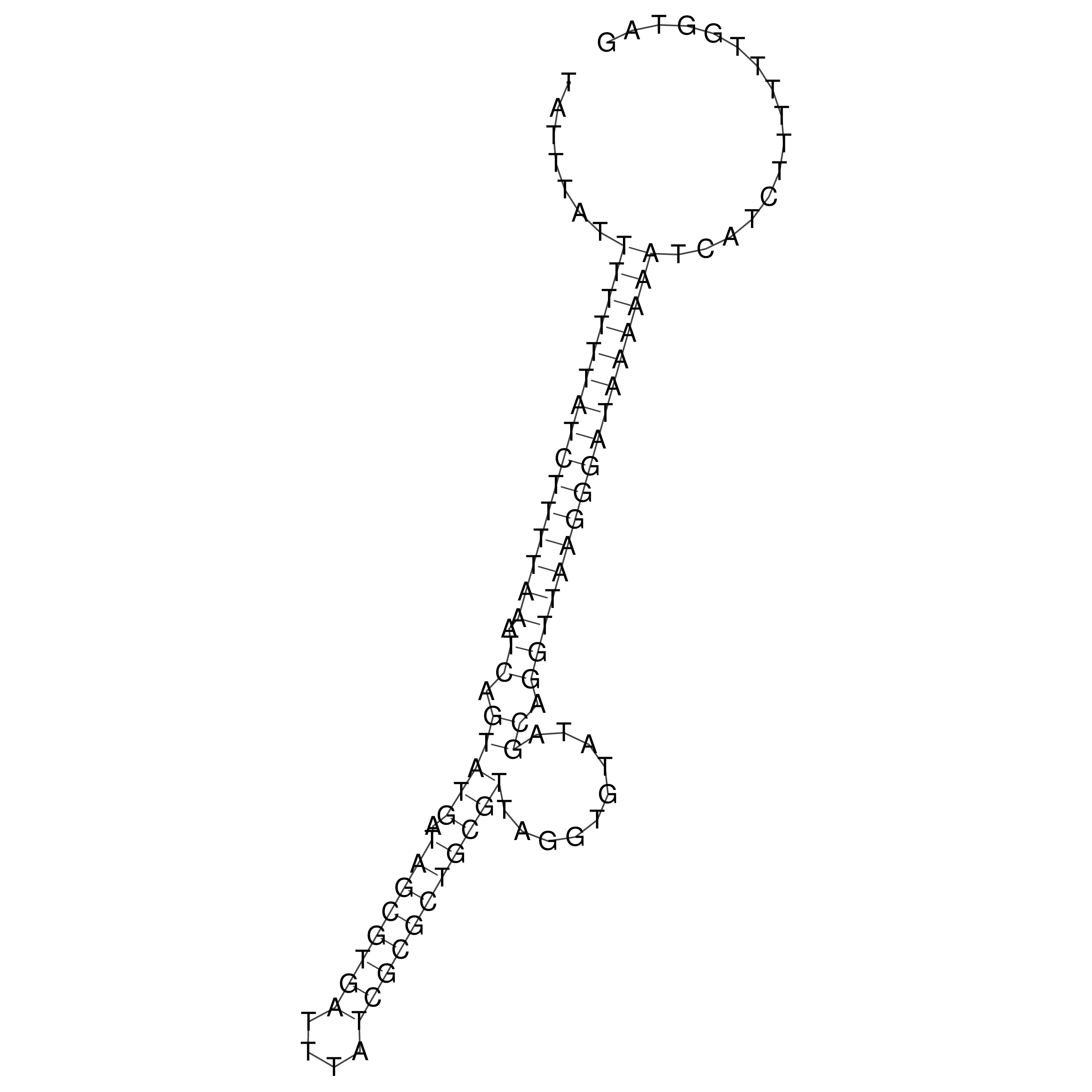
Relaxase
| ID | 3860 | GenBank | WP_001749977 |
| Name | mobF_GVI75_RS27230_pEclE3-3 |
UniProt ID | A0A5C2CVP7 |
| Length | 1080 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1080 a.a. Molecular weight: 120419.59 Da Isoelectric Point: 6.5483
>WP_001749977.1 MULTISPECIES: MobF family relaxase [Gammaproteobacteria]
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLSQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
SSGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEHEEGGHEI
MLDITTITRQNVTSVVGYYSDAKDDYYSKDSSFTSWQGTGAEALGLSGDVESARFKELLVGEIDTFTHMQ
RHVGDAKKERLGYDLTFSAPKGVSMQALIHGDKTIIEAHEKAVAAAVREAEKLAQARTTRQGKSVTQNTN
NLVVATFRHETSRALDPDLHTHAFVMNMTQREDGQWRALKNDELMRNKMHLGDVYKQELALELTKAGYEL
RYNSKNNTFDMAHFSDEQIRAFSRRSEQIEKGLAAMGLTRETADAQTKSRVSMATREKKTEHSREEIHQE
WASRAKTLGIDFDNREWQGHGKPLEADIARNMAPDFTSPEVKADRAIQFAVKSLSERDASFERQKLIQIA
NKQVLGHATIADVEKAYLKAVQKGAIIEGEARYQSTLKVGASVMAETLTRKEWIDSLTNSGMRADKARFA
VDDGIKNGRLKKTSHRVTTVEGIRLERSILTIESRGRGQMPRQLTAEIAGQLLAGKTLKKEQMRAVTEIV
TSKDRFVAAHGYAGTGKSYMTMAAKELLESQGLKVTALAPYGTQKKALEDDGLPARTVAAFLKAKDKKLD
EKSVVFIDEAGVIPARQMKQLMEVIEKHNARAVFLGDTSQTKAVEAGKPFEQLIKAGMQTSYMKDIQRQK
NEVLLEAVKYAAEGNAARALKNITGVNELKEEAPRLSQLADRYLSLSSEQQDATLIISGTNASRKTLNDY
IRGNLGLAGTGETFTLLDRVDSTQAERRDSRYFSKGQIIIPEQDYKNGMKRGESYQVLDTGPGNKLTVES
SSGEQIAFSPRTHTKLSVYQAVSAELAPGDKVMVTRNDKTLDVANGDRFTVKTVEGEKLTLEDKKGRTVE
LDKKQASYLSYAYATTVHKSQGLTCDRVLFNIDTKSLTTSKDVFYVGISRARHEVEIFTDDKKSLASSVS
RDSPKTTAAEIDRFFGLEARFKDIGRDTSLETRSAEKGLPEATGESMAFNQKPDEHNMTTGTDYQPVSNA
EDAFHLKQNPMDDSVGLRRHEAQQNDAELAHDYAAADDQQWSAQEYADYEHYAEASDYDFDSSIYDDYAM
PQTSQAEQSHTGKEHTHEHEHEHEEGGHEI
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5C2CVP7 |
Auxiliary protein
| ID | 1746 | GenBank | WP_001749975 |
| Name | WP_001749975_pEclE3-3 |
UniProt ID | A0A5C2CVS6 |
| Length | 138 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 138 a.a. Molecular weight: 15332.60 Da Isoelectric Point: 5.9670
>WP_001749975.1 MULTISPECIES: hypothetical protein [Gammaproteobacteria]
MPIITAKVSDELLAYIDLVSGGNRSDYLRRCIEAGPGDRESGLKIVADRLSDVNRKLDYLFDRASDADFG
PLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIAEPGKTKAAKAEVERNGYKVWEPKKER
MPIITAKVSDELLAYIDLVSGGNRSDYLRRCIEAGPGDRESGLKIVADRLSDVNRKLDYLFDRASDADFG
PLRDELKAITETLSGVKFPPAGQMMLHESLAIETLILLRSIAEPGKTKAAKAEVERNGYKVWEPKKER
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5C2CVS6 |
T4CP
| ID | 3873 | GenBank | WP_001749976 |
| Name | t4cp2_GVI75_RS27235_pEclE3-3 |
UniProt ID | _ |
| Length | 509 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 509 a.a. Molecular weight: 57679.73 Da Isoelectric Point: 9.5788
>WP_001749976.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Gammaproteobacteria]
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVSARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELRDI
MDDRERGLAFLFAITLPPVMVWFLVAKFTYGIDPSTAKYLIPYLVKNTFSLWPLWSALIAGWFIGVGGLI
AFIIYDKSRVFKGERFKKIYRGTELVSARTLADKTRERGVNQLTVANIPIPTYAENLHFSIAGTTGTGKT
TIFNELLFKSIIRGGKNIALDPNGGFLKNFYRPGDVILNAYDKRTEGWVFFNEIRRSYDYERLVNSIVQE
SPDMATEEWFGYGRLIFSEVSKKLHSLYSTVTMEEVIHWACNVDQKKLKEFLMGTPAEAIFSGSEKAVGS
ARFVLSKNLAPHLKMPEGNFSLRDWLDDGKPGTLFITWQEEMKRSLNPLISCWLDSIFSIVLGMGEKESR
INVFIDELESLQFLPNLNDALTKGRKSGLCVYAGYQTYSQLVKVYGRDMAQTILANMRSNIVLGGSRLGD
ETLDQMSRSLGEIEGEVERKESDPQKPWIVRKRRDVKVVRAVTPTEISMLPNLTGYLALPGDMPVAKFKA
KHVKYHRKNPVPGIELRDI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 21819..41685
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GVI75_RS27095 (GVI75_27090) | 17664..19097 | - | 1434 | WP_001288432 | DNA cytosine methyltransferase | - |
| GVI75_RS27100 (GVI75_27095) | 19196..19468 | + | 273 | Protein_21 | IS1 family transposase | - |
| GVI75_RS27105 (GVI75_27100) | 19479..19685 | - | 207 | WP_001749967 | hypothetical protein | - |
| GVI75_RS27110 (GVI75_27105) | 19690..20337 | - | 648 | WP_015344958 | restriction endonuclease | - |
| GVI75_RS27115 (GVI75_27110) | 20387..20698 | - | 312 | WP_001452736 | hypothetical protein | - |
| GVI75_RS27120 (GVI75_27115) | 20734..21048 | - | 315 | WP_001749965 | TrbM/KikA/MpfK family conjugal transfer protein | - |
| GVI75_RS27125 (GVI75_27120) | 21045..21389 | - | 345 | WP_001749964 | hypothetical protein | - |
| GVI75_RS27130 (GVI75_27125) | 21405..21776 | - | 372 | WP_011867773 | H-NS family nucleoid-associated regulatory protein | - |
| GVI75_RS27135 (GVI75_27130) | 21819..22553 | + | 735 | WP_001749963 | lytic transglycosylase domain-containing protein | virB1 |
| GVI75_RS27140 (GVI75_27135) | 22562..22843 | + | 282 | WP_000440698 | transcriptional repressor KorA | - |
| GVI75_RS27145 (GVI75_27140) | 22853..23146 | + | 294 | WP_001749962 | hypothetical protein | virB2 |
| GVI75_RS27150 (GVI75_27145) | 23196..23513 | + | 318 | WP_000496058 | VirB3 family type IV secretion system protein | virB3 |
| GVI75_RS27155 (GVI75_27150) | 23645..26113 | + | 2469 | WP_224743086 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| GVI75_RS27160 (GVI75_27155) | 26131..26844 | + | 714 | WP_001749960 | type IV secretion system protein | virB5 |
| GVI75_RS27165 (GVI75_27160) | 26852..27079 | + | 228 | WP_001749959 | IncN-type entry exclusion lipoprotein EexN | - |
| GVI75_RS27170 (GVI75_27165) | 27095..28135 | + | 1041 | WP_001749958 | type IV secretion system protein | virB6 |
| GVI75_RS27975 | 28236..28364 | + | 129 | WP_071882546 | conjugal transfer protein TraN | - |
| GVI75_RS27175 (GVI75_27170) | 28354..29052 | + | 699 | WP_000646594 | type IV secretion system protein | virB8 |
| GVI75_RS27180 (GVI75_27175) | 29063..29947 | + | 885 | WP_000735066 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| GVI75_RS27185 (GVI75_27180) | 29947..31107 | + | 1161 | WP_000101710 | type IV secretion system protein VirB10 | virB10 |
| GVI75_RS27190 (GVI75_27185) | 31149..32144 | + | 996 | WP_000128596 | ATPase, T2SS/T4P/T4SS family | virB11 |
| GVI75_RS27195 (GVI75_27190) | 32144..32677 | + | 534 | WP_000792636 | phospholipase D family protein | - |
| GVI75_RS27860 | 32851..33303 | - | 453 | WP_001749956 | DUF6710 family protein | - |
| GVI75_RS27205 (GVI75_27200) | 33614..33874 | + | 261 | WP_197386184 | relaxase | - |
| GVI75_RS27210 (GVI75_27205) | 33906..34556 | - | 651 | WP_000164043 | tetracycline resistance transcriptional repressor TetR(A) | - |
| GVI75_RS27215 (GVI75_27210) | 34662..35861 | + | 1200 | WP_012579085 | tetracycline efflux MFS transporter Tet(A) | - |
| GVI75_RS27220 (GVI75_27215) | 35893..36546 | - | 654 | Protein_46 | EamA family transporter | - |
| GVI75_RS27225 (GVI75_27220) | 36741..36914 | - | 174 | WP_001749978 | hypothetical protein | - |
| GVI75_RS27230 (GVI75_27225) | 36914..40156 | - | 3243 | WP_001749977 | MobF family relaxase | - |
| GVI75_RS27235 (GVI75_27230) | 40156..41685 | - | 1530 | WP_001749976 | type IV secretion system DNA-binding domain-containing protein | virb4 |
| GVI75_RS27240 (GVI75_27235) | 41687..42103 | - | 417 | WP_001749975 | hypothetical protein | - |
| GVI75_RS27865 | 42594..43013 | + | 420 | WP_001749974 | plasmid stabilization protein StbA | - |
| GVI75_RS27250 (GVI75_27245) | 43022..43738 | + | 717 | WP_001749973 | StbB family protein | - |
| GVI75_RS27255 (GVI75_27250) | 43740..44108 | + | 369 | WP_001749972 | plasmid stabilization protein StbC | - |
| GVI75_RS27260 (GVI75_27255) | 44290..44634 | + | 345 | WP_000999874 | hypothetical protein | - |
| GVI75_RS27265 (GVI75_27260) | 44745..45065 | - | 321 | WP_001749971 | protein CcgAII | - |
| GVI75_RS27270 (GVI75_27265) | 45120..45299 | - | 180 | WP_000214483 | protein CcgAI | - |
| GVI75_RS27275 (GVI75_27270) | 46026..46535 | - | 510 | WP_000129958 | antirestriction protein ArdA | - |
Host bacterium
| ID | 6139 | GenBank | NZ_CP049024 |
| Plasmid name | pEclE3-3 | Incompatibility group | IncN |
| Plasmid size | 55421 bp | Coordinate of oriT [Strand] | 42252..42352 [+] |
| Host baterium | Enterobacter hormaechei strain Eho-E3 |
Cargo genes
| Drug resistance gene | aac(6')-Ib-cr, ARR-3, dfrA27, aadA16, qacE, sul1, qnrB6, tet(A) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |