Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105695 |
| Name | oriT_pRIVM_C012369_1 |
| Organism | Enterobacter hormaechei strain RIVM_C012369 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP071024 (201272..201526 [-], 255 nt) |
| oriT length | 255 nt |
| IRs (inverted repeats) | 153..158, 160..165 (AAAAGT..ACTTTT) 122..130, 135..143 (TTAAGGCTT..AAGCCTTAA) 49..55, 60..66 (GAATTTT..AAAATTC) |
| Location of nic site | 78..79 |
| Conserved sequence flanking the nic site |
TTTGGTTAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 255 nt
>oriT_pRIVM_C012369_1
GGATTTAGGTTTTTTTTTAATCGCTTCACATTTCGTTAGCATGCGCAGGAATTTTTGATAAAATTCTGGTTAGTTTGGTTAAAAAGTGTTACAAGTAAGCGGTGTGGTTGAAGGGATAGATTTAAGGCTTATTCAAGCCTTAAGAAAATACTAAAAGTTACTTTTCACCCTACCGAACACCTAACAAAAAATCCATGTTGAAGATTTGAACAATTGTAATGGCGCAAGGACAATCAGCACATGTCAGAATCTGAT
GGATTTAGGTTTTTTTTTAATCGCTTCACATTTCGTTAGCATGCGCAGGAATTTTTGATAAAATTCTGGTTAGTTTGGTTAAAAAGTGTTACAAGTAAGCGGTGTGGTTGAAGGGATAGATTTAAGGCTTATTCAAGCCTTAAGAAAATACTAAAAGTTACTTTTCACCCTACCGAACACCTAACAAAAAATCCATGTTGAAGATTTGAACAATTGTAATGGCGCAAGGACAATCAGCACATGTCAGAATCTGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file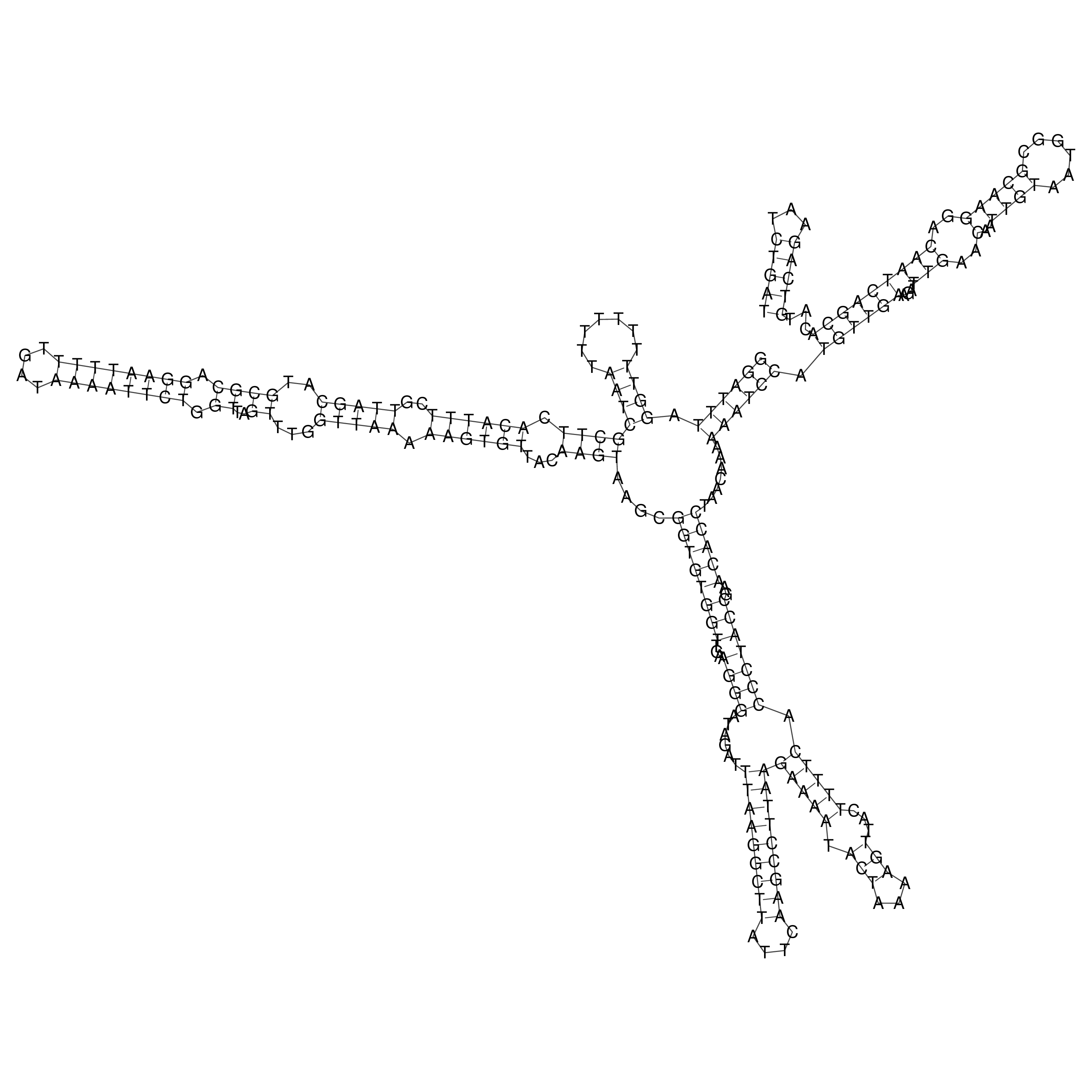
Relaxase
| ID | 3853 | GenBank | WP_012477394 |
| Name | mobH_JQL72_RS23400_pRIVM_C012369_1 |
UniProt ID | A0A2Z2E7B6 |
| Length | 1050 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1050 a.a. Molecular weight: 118067.47 Da Isoelectric Point: 4.6347
>WP_012477394.1 MULTISPECIES: MobH family relaxase [Enterobacterales]
MMMNFRALYLCIKRILGIFSSQENDATSVMIEDISSLSPFAQILGDQKYTVPDHPNPEVLKFIEYPTRPT
GIQTFNEQSILSLYREKLHSISMMLAISDSDIRDDAYTFTNLVLKPLVEYVRWIHLLPASENHHHNGIGG
LLSHSLEVAILSLKNAHHSELRPIGYQDEEVVRRKVYLYAAFICGLVHDAGKVYDLDIVSLNLASPIIWT
PSSQSLLDWARENDVVEYEIHWRKRIHNQHNIWSSVFLERILNPVCLAFLDRVNKERVYSKMITALNVYT
DGNDFLSKCVRTADFYSTGTDLNVLRDPIMGLRSNDAAARAISTIKHNFTSININNYNAKPMHIIIVNGE
VYLNENAFLDFVLNDFELHKYNFPQGEAGKTVLVESLVQRGYVEPYDDERVVHYFIPGIYSENEISNIFR
NGIGKLEFYNLLKLRWIGLIFDSYKIPDSVPGLFSVNANKDFIYIDEQKTVTEYRRPVPGRDVITKITDT
VETAVLKVNDLGRSSASIDVDIHSKKNEGSSDDFEKKAESDNEIDNDTQIVKSEGEEAADPVIPDIEESE
DESAKDTESHVLVNQLHELLLSAPLSNDYIVCVDAVPYLNIDTTMALLPGLDEKAFSEEPYFQLTFREGS
LDGMWIVRDIDDLRLVQLGDNCAGFQLTYHEPRRPTTLKSLFNTSMYQALVINDESSVENSAPRPKQTLE
LPPPRVNAVEEHSGDVEYHGTDSASATGPLKTEAVEYEHYQHLFEKEDEEHEIIDYTDFSQLSVSRPEVG
SCATSSSVHNEKLLSEPSELPELNREQNADPQGTNERSMDVSVGQENSEPDTEGNCPPPAEVVYSQTEAA
ATSVMASEEPALPPVLEESNGEHAPTDAKGHHLSPALARLFAPTAPVEKQNPKRNRNKSSDKAEVQKPAS
PVSGHNLNSKVFASTESDQNGEFSLISEGDVTELEFVEIALVLHQILSKMEVAFKRKRKNRFMVSTPNTL
YLTQSCVEKFGSQLEAQDLFNKLPQYLVNSGAVINTKCHAFNMPTLLAASDRAKVDIERIINNLKEAGNL
MMMNFRALYLCIKRILGIFSSQENDATSVMIEDISSLSPFAQILGDQKYTVPDHPNPEVLKFIEYPTRPT
GIQTFNEQSILSLYREKLHSISMMLAISDSDIRDDAYTFTNLVLKPLVEYVRWIHLLPASENHHHNGIGG
LLSHSLEVAILSLKNAHHSELRPIGYQDEEVVRRKVYLYAAFICGLVHDAGKVYDLDIVSLNLASPIIWT
PSSQSLLDWARENDVVEYEIHWRKRIHNQHNIWSSVFLERILNPVCLAFLDRVNKERVYSKMITALNVYT
DGNDFLSKCVRTADFYSTGTDLNVLRDPIMGLRSNDAAARAISTIKHNFTSININNYNAKPMHIIIVNGE
VYLNENAFLDFVLNDFELHKYNFPQGEAGKTVLVESLVQRGYVEPYDDERVVHYFIPGIYSENEISNIFR
NGIGKLEFYNLLKLRWIGLIFDSYKIPDSVPGLFSVNANKDFIYIDEQKTVTEYRRPVPGRDVITKITDT
VETAVLKVNDLGRSSASIDVDIHSKKNEGSSDDFEKKAESDNEIDNDTQIVKSEGEEAADPVIPDIEESE
DESAKDTESHVLVNQLHELLLSAPLSNDYIVCVDAVPYLNIDTTMALLPGLDEKAFSEEPYFQLTFREGS
LDGMWIVRDIDDLRLVQLGDNCAGFQLTYHEPRRPTTLKSLFNTSMYQALVINDESSVENSAPRPKQTLE
LPPPRVNAVEEHSGDVEYHGTDSASATGPLKTEAVEYEHYQHLFEKEDEEHEIIDYTDFSQLSVSRPEVG
SCATSSSVHNEKLLSEPSELPELNREQNADPQGTNERSMDVSVGQENSEPDTEGNCPPPAEVVYSQTEAA
ATSVMASEEPALPPVLEESNGEHAPTDAKGHHLSPALARLFAPTAPVEKQNPKRNRNKSSDKAEVQKPAS
PVSGHNLNSKVFASTESDQNGEFSLISEGDVTELEFVEIALVLHQILSKMEVAFKRKRKNRFMVSTPNTL
YLTQSCVEKFGSQLEAQDLFNKLPQYLVNSGAVINTKCHAFNMPTLLAASDRAKVDIERIINNLKEAGNL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A2Z2E7B6 |
T4CP
| ID | 3862 | GenBank | WP_241102379 |
| Name | t4cp1_JQL72_RS23405_pRIVM_C012369_1 |
UniProt ID | _ |
| Length | 344 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 344 a.a. Molecular weight: 39355.54 Da Isoelectric Point: 8.8550
>WP_241102379.1 ATP-binding protein [Enterobacter hormaechei]
MSDSKRTNLHAQENFYRPILEYRSASILLICSVSMLYMGLSSDGLDIAPIVLFTSILLFLLCLYRCKTAA
PFLMAHWRVFKRHFMFVSLDSLRVINKSNFFSNERKYRQLVQDYQNKNKDIPERKSYFCDGFEWGPEHAD
RAYQIANLSSDKREIELPFVFNPIKRHFDAMARKMGGSNAIFAVERREPIFVTEDNWFGHTLITGNVGTG
KTVLQRLLSISMLHLGHVVVVIDPKNDAEWRESLMEEAKTLGLPFYKFHPGQPASSVCIDVCNTYTNVSD
LTSRLLSLVTVPGEVNPFVQYAKALVSNVISGLSYIEKKPSIYLIHKNMKSHIDLPPRLDTTLS
MSDSKRTNLHAQENFYRPILEYRSASILLICSVSMLYMGLSSDGLDIAPIVLFTSILLFLLCLYRCKTAA
PFLMAHWRVFKRHFMFVSLDSLRVINKSNFFSNERKYRQLVQDYQNKNKDIPERKSYFCDGFEWGPEHAD
RAYQIANLSSDKREIELPFVFNPIKRHFDAMARKMGGSNAIFAVERREPIFVTEDNWFGHTLITGNVGTG
KTVLQRLLSISMLHLGHVVVVIDPKNDAEWRESLMEEAKTLGLPFYKFHPGQPASSVCIDVCNTYTNVSD
LTSRLLSLVTVPGEVNPFVQYAKALVSNVISGLSYIEKKPSIYLIHKNMKSHIDLPPRLDTTLS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3863 | GenBank | WP_236904384 |
| Name | t4cp2_JQL72_RS23415_pRIVM_C012369_1 |
UniProt ID | _ |
| Length | 362 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 362 a.a. Molecular weight: 40108.90 Da Isoelectric Point: 6.2444
>WP_236904384.1 MULTISPECIES: type IV secretion system DNA-binding domain-containing protein [Enterobacter cloacae complex]
MSIVNLTVKVMESCYARYYGYDVWTEKVKYVANDTLPVRFKRLAEWFTAHFMNYEGSEQIDWLDTVSQLI
DYSMSDPEHMAKMTAGIMPVFDMLIEKPLNELLSPNPNSVSSREIVTSEGMFSTGGVLYISLDGLSNPDT
AAAISQLIMSDLTSCAGSRYNAQDGDMSANSRISIFVDEAHSAINNPMINLLAQGRAAKIALFICTQTIS
DFIAAASVETANRITGLCNNYISLRVNDTPTQTLVVENFGKSAISTNMVTYTTGSETSLPHNNFSGSISE
RKQTTLEESIPKDLLGQVPMFHIVARLQDGRKVVGQIPIAVAEKQMKPNTTLSEMLFKKAGKVTLRQNLD
IKNLNKFLRKLH
MSIVNLTVKVMESCYARYYGYDVWTEKVKYVANDTLPVRFKRLAEWFTAHFMNYEGSEQIDWLDTVSQLI
DYSMSDPEHMAKMTAGIMPVFDMLIEKPLNELLSPNPNSVSSREIVTSEGMFSTGGVLYISLDGLSNPDT
AAAISQLIMSDLTSCAGSRYNAQDGDMSANSRISIFVDEAHSAINNPMINLLAQGRAAKIALFICTQTIS
DFIAAASVETANRITGLCNNYISLRVNDTPTQTLVVENFGKSAISTNMVTYTTGSETSLPHNNFSGSISE
RKQTTLEESIPKDLLGQVPMFHIVARLQDGRKVVGQIPIAVAEKQMKPNTTLSEMLFKKAGKVTLRQNLD
IKNLNKFLRKLH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 2429..11636
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JQL72_RS22390 (JQL72_22390) | 1..876 | + | 876 | WP_000594611 | RepB family plasmid replication initiator protein | - |
| JQL72_RS22395 (JQL72_22395) | 2025..2378 | + | 354 | WP_000423602 | hypothetical protein | - |
| JQL72_RS22400 (JQL72_22400) | 2396..2746 | + | 351 | WP_001447770 | type IV conjugative transfer system protein TraL | - |
| JQL72_RS22405 (JQL72_22405) | 2758..3546 | + | 789 | WP_000783153 | TraE/TraK family type IV conjugative transfer system protein | traE |
| JQL72_RS22410 (JQL72_22410) | 3546..4817 | + | 1272 | WP_000592092 | type-F conjugative transfer system secretin TraK | traK |
| JQL72_RS22415 (JQL72_22415) | 4819..5280 | + | 462 | WP_021242895 | plasmid transfer protein HtdO | - |
| JQL72_RS22420 (JQL72_22420) | 5270..6625 | + | 1356 | WP_000351841 | IncHI-type conjugal transfer protein TrhB | traB |
| JQL72_RS22425 (JQL72_22425) | 6633..7115 | + | 483 | WP_000377632 | hypothetical protein | - |
| JQL72_RS22430 (JQL72_22430) | 7234..7986 | + | 753 | WP_182911723 | protein-disulfide isomerase HtdT | - |
| JQL72_RS22435 (JQL72_22435) | 7996..8946 | + | 951 | WP_001022588 | IncHI-type conjugal transfer lipoprotein TrhV | traV |
| JQL72_RS22440 (JQL72_22440) | 8955..11636 | + | 2682 | WP_000387412 | TraC family protein | virb4 |
| JQL72_RS22445 (JQL72_22445) | 14185..14346 | + | 162 | WP_001371949 | hypothetical protein | - |
Host bacterium
| ID | 6132 | GenBank | NZ_CP071024 |
| Plasmid name | pRIVM_C012369_1 | Incompatibility group | IncHI2A |
| Plasmid size | 316078 bp | Coordinate of oriT [Strand] | 201272..201526 [-] |
| Host baterium | Enterobacter hormaechei strain RIVM_C012369 |
Cargo genes
| Drug resistance gene | blaACC-1, msr(E), mph(E), mcr-9, sul1, qacE, ant(3'')-Ia, blaVIM-1 |
| Virulence gene | - |
| Metal resistance gene | terW, terZ, terA, terB, terC, terD, terE, silE, silS, silR, silC, silF, silB, silA, silP, pcoS, pcoE, ncrC, rcnR/yohL, arsC, arsB, arsH, fecD, fecE, arsR, merR, merT, merP, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |