Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105644 |
| Name | oriT_pH29-46 |
| Organism | Mammaliicoccus lentus strain H29 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP059680 (46102..46137 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACG..CGTTCGC) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pH29-46
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file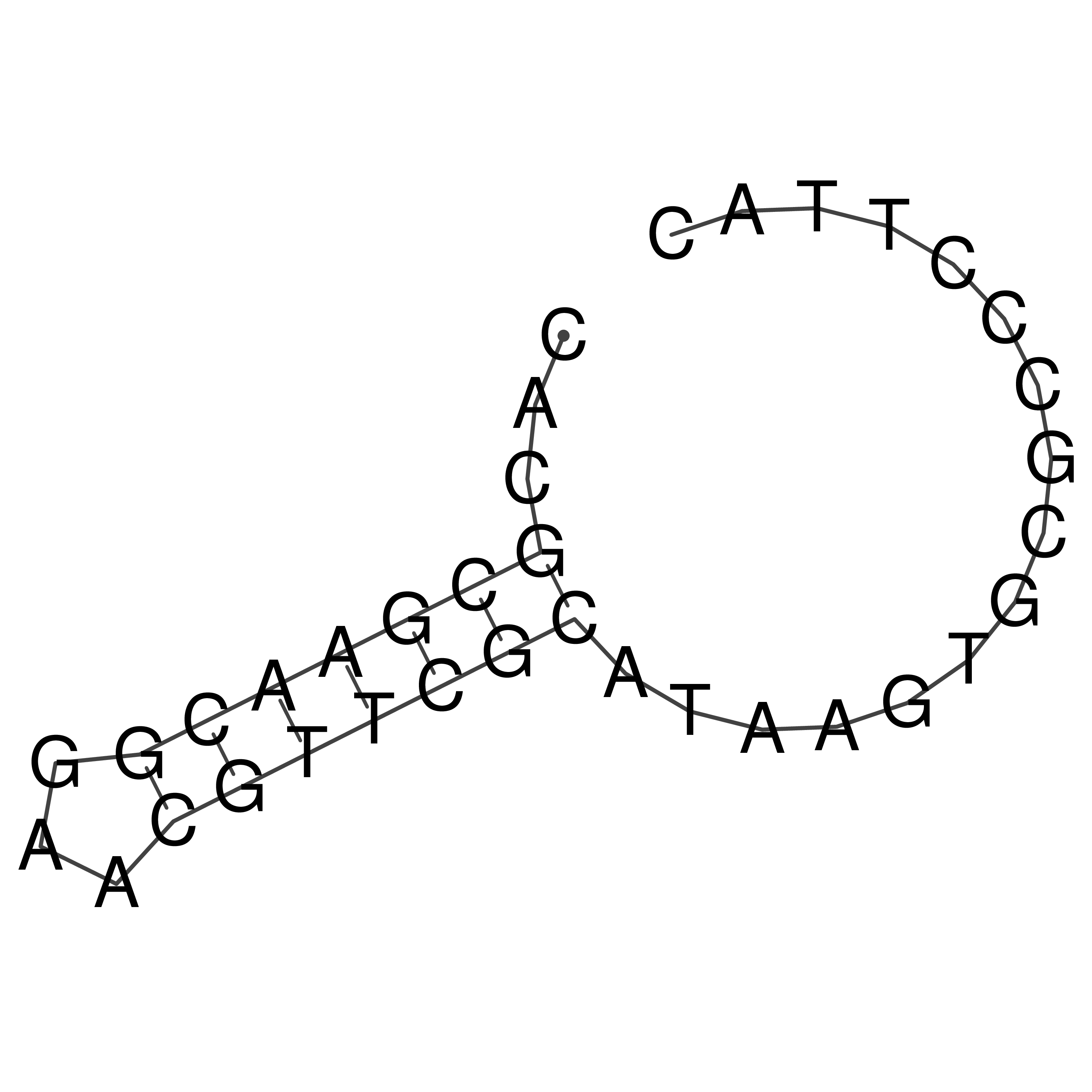
Relaxase
| ID | 3828 | GenBank | WP_182094133 |
| Name | MobA_MobL_H3V22_RS14205_pH29-46 |
UniProt ID | _ |
| Length | 665 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 78741.64 Da Isoelectric Point: 5.1613
>WP_182094133.1 MULTISPECIES: MobA/MobL family protein [Staphylococcaceae]
MAMYHFQNKFVSKANGQSATAKAAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVNDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINDENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKDKHNEWRKRWETVQNKHLEMNGFSVRVSADSYKKQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKINKMFDEVDDMKSHKLNAFSYMDKSDSTTLKDMAKDLKIYV
TPINMYEENERLYDLKQKTALITDDEDRLNKIENIEDRQKKLESINEVFEKQAGIFFDKNYLEQALNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTVIGNRDISIESIEKESNFFADKLSNILDK
NNLSFDDVLENKHEGIEDSLKIDYYTSKLEVLRNAENTLEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTLEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLNEY
DFDYSDNNDLKHLYQESNEVGDETSKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLVNAGLDSMIFNFN
EIFRERMPKYINNQYKGKNHSKQRHELKNKRGMHL
MAMYHFQNKFVSKANGQSATAKAAYNSASRIKDFKENEFKDYSNKQCDYSEILLPNNADDKFKDREYLWN
KVNDVENRKNSQVAREIIIGLPNEFDPNSNIELAKEFAESLSNEGMIVDLNIHKINDENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKDKHNEWRKRWETVQNKHLEMNGFSVRVSADSYKKQNIDLEPTKKEGW
KARKFEDETGKKSSISKHNESIKKRNQQKINKMFDEVDDMKSHKLNAFSYMDKSDSTTLKDMAKDLKIYV
TPINMYEENERLYDLKQKTALITDDEDRLNKIENIEDRQKKLESINEVFEKQAGIFFDKNYLEQALNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTVIGNRDISIESIEKESNFFADKLSNILDK
NNLSFDDVLENKHEGIEDSLKIDYYTSKLEVLRNAENTLEDYYDVQIKELFTDDEDYKAFNEVTDIKEKQ
QLIDFKTYHGTENTLEMLETGNFIPKYSDEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLNEY
DFDYSDNNDLKHLYQESNEVGDETSKDNIEEFYEGNDILVDKETYNNYNRKQQAYGLVNAGLDSMIFNFN
EIFRERMPKYINNQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3836 | GenBank | WP_030061486 |
| Name | t4cp2_H3V22_RS14060_pH29-46 |
UniProt ID | _ |
| Length | 548 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 548 a.a. Molecular weight: 62899.75 Da Isoelectric Point: 5.9714
>WP_030061486.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Staphylococcaceae]
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSFKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKMNKDVWYKSSVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNHDDEGSVELDNRFNELPKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKIKQQKLEEYKEEQ
RQKELEEEQQAQEEQQNEEVKESKSKNEEQQEDKKDEQQKINDSKKAFYEKLKQKQAK
MTTLLGELESKATSINKKNLVIQDFDTKCPNRNIFVVGGPGSFKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKMNKDVWYKSSVGLL
NSLLLYAKYEFEPEKRTIGNIIKFIQNNKPNHDDEGSVELDNRFNELPKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ENGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKEELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMQQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETYIAPQNEYKRKKNQSMIDKHNRLREEWDKKQEKIKQQKLEEYKEEQ
RQKELEEEQQAQEEQQNEEVKESKSKNEEQQEDKKDEQQKINDSKKAFYEKLKQKQAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 5364..16967
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| H3V22_RS13980 (H3V22_13980) | 434..667 | + | 234 | WP_000602212 | hypothetical protein | - |
| H3V22_RS13985 (H3V22_13985) | 930..1970 | + | 1041 | WP_000073390 | hypothetical protein | - |
| H3V22_RS13990 (H3V22_13990) | 1973..2302 | + | 330 | WP_030061459 | plasmid segregation protein ParR | - |
| H3V22_RS13995 (H3V22_13995) | 2306..2566 | - | 261 | WP_001008214 | helix-turn-helix transcriptional regulator | - |
| H3V22_RS14000 (H3V22_14000) | 2949..3908 | + | 960 | WP_030061463 | replication initiator protein A | - |
| H3V22_RS14005 (H3V22_14005) | 4012..4200 | - | 189 | WP_030061465 | hypothetical protein | - |
| H3V22_RS14010 (H3V22_14010) | 4373..5347 | + | 975 | WP_030061467 | conjugative transfer protein TrsA | - |
| H3V22_RS14015 (H3V22_14015) | 5364..5681 | + | 318 | WP_030061469 | CagC family type IV secretion system protein | virB2 |
| H3V22_RS14020 (H3V22_14020) | 5740..6147 | + | 408 | WP_030061471 | hypothetical protein | trsC |
| H3V22_RS14025 (H3V22_14025) | 6134..6817 | + | 684 | WP_030061473 | TrsD/TraD family conjugative transfer protein | trsD |
| H3V22_RS14030 (H3V22_14030) | 6838..8850 | + | 2013 | WP_030061475 | DUF87 domain-containing protein | virb4 |
| H3V22_RS14035 (H3V22_14035) | 8862..10142 | + | 1281 | WP_030061477 | hypothetical protein | trsF |
| H3V22_RS14040 (H3V22_14040) | 10160..11236 | + | 1077 | WP_031921304 | CHAP domain-containing protein | trsG |
| H3V22_RS14045 (H3V22_14045) | 11246..11731 | + | 486 | WP_000735570 | TrsH/TraH family protein | - |
| H3V22_RS14050 (H3V22_14050) | 11747..13849 | + | 2103 | WP_030061483 | DNA topoisomerase III | - |
| H3V22_RS14055 (H3V22_14055) | 13865..14329 | + | 465 | WP_000715271 | hypothetical protein | trsJ |
| H3V22_RS14060 (H3V22_14060) | 14326..15972 | + | 1647 | WP_030061486 | VirD4-like conjugal transfer protein, CD1115 family | - |
| H3V22_RS14065 (H3V22_14065) | 16050..16967 | + | 918 | WP_030061488 | conjugal transfer protein TrbL family protein | trsL |
| H3V22_RS14070 (H3V22_14070) | 16984..17379 | + | 396 | WP_000608969 | single-stranded DNA-binding protein | - |
| H3V22_RS14075 (H3V22_14075) | 17436..18167 | + | 732 | WP_000869299 | hypothetical protein | - |
| H3V22_RS14080 (H3V22_14080) | 18204..18827 | + | 624 | WP_030061493 | hypothetical protein | - |
| H3V22_RS14085 (H3V22_14085) | 18843..19460 | + | 618 | WP_182094135 | hypothetical protein | - |
| H3V22_RS14090 (H3V22_14090) | 19588..20672 | + | 1085 | Protein_23 | tyrosine-type recombinase/integrase | - |
Host bacterium
| ID | 6081 | GenBank | NZ_CP059680 |
| Plasmid name | pH29-46 | Incompatibility group | - |
| Plasmid size | 46167 bp | Coordinate of oriT [Strand] | 46102..46137 [+] |
| Host baterium | Mammaliicoccus lentus strain H29 |
Cargo genes
| Drug resistance gene | fexA, cfr, aac(6')-aph(2'') |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |