Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105618 |
| Name | oriT_pRetNXC12e |
| Organism | Rhizobium etli strain NXC12 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP020911 (284291..284319 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pRetNXC12e
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file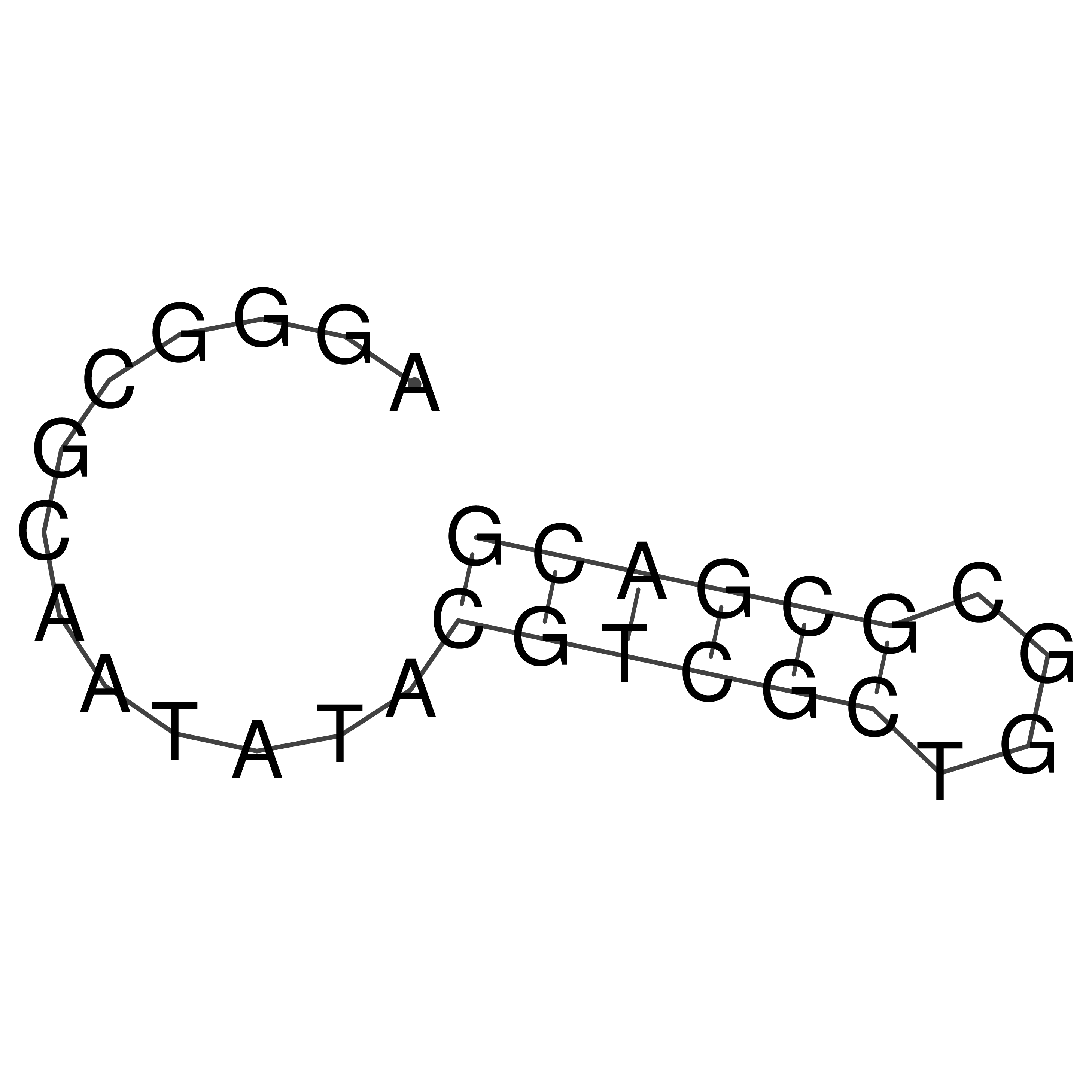
Relaxase
| ID | 3814 | GenBank | WP_086084174 |
| Name | traA_NXC12_RS29575_pRetNXC12e |
UniProt ID | _ |
| Length | 1543 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1543 a.a. Molecular weight: 171228.51 Da Isoelectric Point: 9.0655
>WP_086084174.1 Ti-type conjugative transfer relaxase TraA [Rhizobium etli]
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGAGELMHEELALPDEIPDWLRSAISG
QSVSNASEVFWNAVDAFETRGDAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEDGFGRKKVPVLGGDGKPLRVITPDRPNGNIVYKVWAGDKETLKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQTHLGPEKAALARKGRELHFAPADLARRQEMADRLLADPERLLTQ
LVNERSTFDERDIAKALHRYVDDPSDFANIRARLMASTELVMLKPQEIDTQTGKASEPAVFTTRDMLRIE
YEMAQSARVLSGRRDFGVSERHVSAAIESIETRDPQKRFRLDPEQVDAIRHVTGDDGIAAVVGLAGAGKS
TLLAAARLAWESQGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELAWANGRDTLHRGDVVVIDEAGMVS
SPQMARVLDIVEQAEAKVVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQRHGWARDASRLFARGE
IDKGLDAYAGHGHLVEAETRQEIVDRIVADWREARRQAIGRSVSEGRDGSLGGDELLVLAHTNADVKRLN
EALRSVMTGEGALGESRSFRTERGAREFAAGDRVIFLENARFIEPRARRLGPQYVKNGMLGTVVSTGDAK
GEVLLSVRLDNGSKVVFGEESYRNVDHGYAATIHKSQGATVERTFVLASGMMDRHLTYVSMTRHRDRADL
YAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADLRTGDGTVHRLWGVSLPKA
LEDAGILEGDTVTLRKDGIERVKVEVPVIGEKTGQKRYEEREVDRNVWTAKQIETATERQERIERESHRP
DVFKQLVERLSRSGAKTTTLDFDSEAGYRAHAQDFARRRGIEHLSLVAAEMEESLSRRWAWIAEKREQVE
KLWERASVALGFAIERERRISYNEQRAETRAEVRSLYTRYLIQPATAFARSVDEDARLAQLSSPAWKEQE
AILRPVLEKIYRDPDAALVALNALASNTSIAPRELASNLAAAPDRLGRLRGSDLIVDGRAARDERNVAMA
ALQELLPLARAHATGFRRNAERFELREQTRRAHMSLSIPALSERAMTRLMEIEAVRAQGGDDAYKMAFAL
AAQDRTVVREIKAVSEAITARFGWSAFTAKADAIADRNMVDRMPEDLTAGRREELTPLFEAVKRFAEEQH
LAERQDRSKIVAGASADLSKEPGKEKVTMPPTFAAVTEFKVLVEEEARSRALASPLYRQQRAAMADAART
IWRDPAGAVGRIEELLSKGFAADRIAAAVTNDPAAYGPLRGSSRLMDRMLASGRERKAAIEAVPETAARL
RSFGAAYANIFDAGRHAITEERRRMAVAIPALSKSAEEALTRLTAEARKDSRRLSVSAASLDPNIRREFA
AVSRALDDRFGRNAIVRGDKDLINRVPPPQRRVFEAMQERLKVLQQTVRLQSSEQIIVERRQRTSSRARG
INL
MAIMFVRAQVISRGAGRSIVSAAAYRHRTRMMDEQAGTSFSYRGGAGELMHEELALPDEIPDWLRSAISG
QSVSNASEVFWNAVDAFETRGDAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMIADWVYHDKDG
NPHIHLMTTLRPLTEDGFGRKKVPVLGGDGKPLRVITPDRPNGNIVYKVWAGDKETLKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQTHLGPEKAALARKGRELHFAPADLARRQEMADRLLADPERLLTQ
LVNERSTFDERDIAKALHRYVDDPSDFANIRARLMASTELVMLKPQEIDTQTGKASEPAVFTTRDMLRIE
YEMAQSARVLSGRRDFGVSERHVSAAIESIETRDPQKRFRLDPEQVDAIRHVTGDDGIAAVVGLAGAGKS
TLLAAARLAWESQGRRVIGAALAGKAAEGLEDSSGIKSRTLASWELAWANGRDTLHRGDVVVIDEAGMVS
SPQMARVLDIVEQAEAKVVLVGDAMQLQPIQAGAAFRAISERIGFAELAGVRRQRHGWARDASRLFARGE
IDKGLDAYAGHGHLVEAETRQEIVDRIVADWREARRQAIGRSVSEGRDGSLGGDELLVLAHTNADVKRLN
EALRSVMTGEGALGESRSFRTERGAREFAAGDRVIFLENARFIEPRARRLGPQYVKNGMLGTVVSTGDAK
GEVLLSVRLDNGSKVVFGEESYRNVDHGYAATIHKSQGATVERTFVLASGMMDRHLTYVSMTRHRDRADL
YAAKEDFAAKPEWGRKPRVDHAAGVTGELVETGEAKFRPDDEDADDSPYADLRTGDGTVHRLWGVSLPKA
LEDAGILEGDTVTLRKDGIERVKVEVPVIGEKTGQKRYEEREVDRNVWTAKQIETATERQERIERESHRP
DVFKQLVERLSRSGAKTTTLDFDSEAGYRAHAQDFARRRGIEHLSLVAAEMEESLSRRWAWIAEKREQVE
KLWERASVALGFAIERERRISYNEQRAETRAEVRSLYTRYLIQPATAFARSVDEDARLAQLSSPAWKEQE
AILRPVLEKIYRDPDAALVALNALASNTSIAPRELASNLAAAPDRLGRLRGSDLIVDGRAARDERNVAMA
ALQELLPLARAHATGFRRNAERFELREQTRRAHMSLSIPALSERAMTRLMEIEAVRAQGGDDAYKMAFAL
AAQDRTVVREIKAVSEAITARFGWSAFTAKADAIADRNMVDRMPEDLTAGRREELTPLFEAVKRFAEEQH
LAERQDRSKIVAGASADLSKEPGKEKVTMPPTFAAVTEFKVLVEEEARSRALASPLYRQQRAAMADAART
IWRDPAGAVGRIEELLSKGFAADRIAAAVTNDPAAYGPLRGSSRLMDRMLASGRERKAAIEAVPETAARL
RSFGAAYANIFDAGRHAITEERRRMAVAIPALSKSAEEALTRLTAEARKDSRRLSVSAASLDPNIRREFA
AVSRALDDRFGRNAIVRGDKDLINRVPPPQRRVFEAMQERLKVLQQTVRLQSSEQIIVERRQRTSSRARG
INL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3819 | GenBank | WP_086084177 |
| Name | traG_NXC12_RS29590_pRetNXC12e |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70326.36 Da Isoelectric Point: 9.4854
>WP_086084177.1 Ti-type conjugative transfer system protein TraG [Rhizobium etli]
MGLRGKPHPSLLLVLVPIAVTTITIYVVGWRWPELAAGMSGKIGYSFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLCFLGVAAFYALREFGRLAPAVQARVVPWDRALSYVDMVAVIAAVAGFLAVAVSARI
SVVVPDEIKRARRGIFGDADWLPMAAAGKLFPPDGEIVVGERYRVDREIVHALPFDANDRSTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALKYTGPLICLDPSTEVAPIVVEHRTNALRREVMVLDPT
NPTMGFNVLDGIETSSQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLATLRDIQENSQADFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRTLFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAIKALDTARSVSAQCGEMTVEVKG
SSRNLGWDKRAAGPRKSESISFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKEMDAVAK
ANRFVKSIT
MGLRGKPHPSLLLVLVPIAVTTITIYVVGWRWPELAAGMSGKIGYSFLRAAPVPPLLFGPLAGLLTVWAL
PLHRRRPVAMASLLCFLGVAAFYALREFGRLAPAVQARVVPWDRALSYVDMVAVIAAVAGFLAVAVSARI
SVVVPDEIKRARRGIFGDADWLPMAAAGKLFPPDGEIVVGERYRVDREIVHALPFDANDRSTWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVVPTALKYTGPLICLDPSTEVAPIVVEHRTNALRREVMVLDPT
NPTMGFNVLDGIETSSQKEEDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLREIVSEPEPSVLATLRDIQENSQADFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKPSDIVSGRKDVFLNIPASILRSYPGIGRVIIGSLINAMMQADGAFRRRTLFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIEGCAFASYAAIKALDTARSVSAQCGEMTVEVKG
SSRNLGWDKRAAGPRKSESISFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRCGRAIYFRRKEMDAVAK
ANRFVKSIT
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 292877..307414
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| NXC12_RS33705 (NXC12_PE00263) | 288556..288723 | - | 168 | WP_016735266 | hypothetical protein | - |
| NXC12_RS29620 (NXC12_PE00264) | 289038..289361 | - | 324 | WP_008530485 | DUF736 family protein | - |
| NXC12_RS29625 (NXC12_PE00265) | 289824..290156 | + | 333 | WP_198388454 | WGR domain-containing protein | - |
| NXC12_RS29630 (NXC12_PE00266) | 290291..290692 | - | 402 | WP_086084182 | type II toxin-antitoxin system VapC family toxin | - |
| NXC12_RS29635 (NXC12_PE00267) | 290692..290928 | - | 237 | WP_086084183 | type II toxin-antitoxin system VapB family antitoxin | - |
| NXC12_RS29640 (NXC12_PE00268) | 291516..292724 | + | 1209 | WP_086084184 | DUF1173 domain-containing protein | - |
| NXC12_RS29645 (NXC12_PE00269) | 292877..293908 | - | 1032 | WP_086084185 | P-type DNA transfer ATPase VirB11 | virB11 |
| NXC12_RS29650 (NXC12_PE00270) | 293917..295092 | - | 1176 | WP_086084186 | type IV secretion system protein VirB10 | virB10 |
| NXC12_RS29655 (NXC12_PE00271) | 295101..295958 | - | 858 | WP_086084187 | P-type conjugative transfer protein VirB9 | virB9 |
| NXC12_RS29660 (NXC12_PE00272) | 295955..296626 | - | 672 | WP_086084188 | virB8 family protein | virB8 |
| NXC12_RS34310 (NXC12_PE00273) | 296628..296912 | - | 285 | WP_086084189 | hypothetical protein | - |
| NXC12_RS29670 (NXC12_PE00274) | 296947..297879 | - | 933 | WP_198388457 | type IV secretion system protein | virB6 |
| NXC12_RS29675 (NXC12_PE00275) | 297882..298115 | - | 234 | WP_086084190 | EexN family lipoprotein | - |
| NXC12_RS29680 (NXC12_PE00276) | 298112..298813 | - | 702 | WP_157700475 | P-type DNA transfer protein VirB5 | virB5 |
| NXC12_RS29685 (NXC12_PE00277) | 298810..301176 | - | 2367 | WP_086084191 | VirB4 family type IV secretion system protein | virb4 |
| NXC12_RS29690 (NXC12_PE00278) | 301169..301507 | - | 339 | WP_086084192 | type IV secretion system protein VirB3 | virB3 |
| NXC12_RS29695 (NXC12_PE00279) | 301513..301812 | - | 300 | WP_086084193 | TrbC/VirB2 family protein | virB2 |
| NXC12_RS29700 (NXC12_PE00280) | 301809..302468 | - | 660 | WP_086084194 | lytic transglycosylase domain-containing protein | virB1 |
| NXC12_RS29705 (NXC12_PE00281) | 302472..303008 | - | 537 | WP_086084195 | hypothetical protein | - |
| NXC12_RS29710 (NXC12_PE00282) | 303107..303472 | + | 366 | WP_020919765 | MarR family transcriptional regulator | - |
| NXC12_RS29715 | 303825..304107 | + | 283 | Protein_290 | HU family DNA-binding protein | - |
| NXC12_RS34315 (NXC12_PE00284) | 304162..304326 | - | 165 | WP_244920145 | hypothetical protein | - |
| NXC12_RS33350 | 304736..305134 | + | 399 | WP_157700476 | hypothetical protein | - |
| NXC12_RS29730 (NXC12_PE00286) | 305131..306066 | - | 936 | WP_086084517 | DUF2493 domain-containing protein | - |
| NXC12_RS29735 (NXC12_PE00287) | 306380..307414 | - | 1035 | WP_020919760 | toprim domain-containing protein | traP |
| NXC12_RS34655 (NXC12_PE00288) | 307427..308200 | - | 774 | WP_020919759 | DUF6117 family protein | - |
| NXC12_RS29745 (NXC12_PE00289) | 308289..309479 | - | 1191 | WP_020919758 | hypothetical protein | - |
| NXC12_RS29750 (NXC12_PE00290) | 309556..310212 | - | 657 | WP_020919757 | hypothetical protein | - |
| NXC12_RS29755 (NXC12_PE00291) | 310285..312402 | - | 2118 | WP_020919756 | ParB/RepB/Spo0J family partition protein | - |
Host bacterium
| ID | 6056 | GenBank | NZ_CP020911 |
| Plasmid name | pRetNXC12e | Incompatibility group | - |
| Plasmid size | 978479 bp | Coordinate of oriT [Strand] | 284291..284319 [-] |
| Host baterium | Rhizobium etli strain NXC12 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bphB, LinG, dxnH, bphC |
| Symbiosis gene | fixB, fixA, fixG, fixQ, fixO, fixN |
| Anti-CRISPR | AcrIIA9 |