Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105567 |
| Name | oriT_bat15|unnamed2 |
| Organism | Klebsiella pneumoniae strain bat15 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP092767 (94507..94556 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_bat15|unnamed2
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file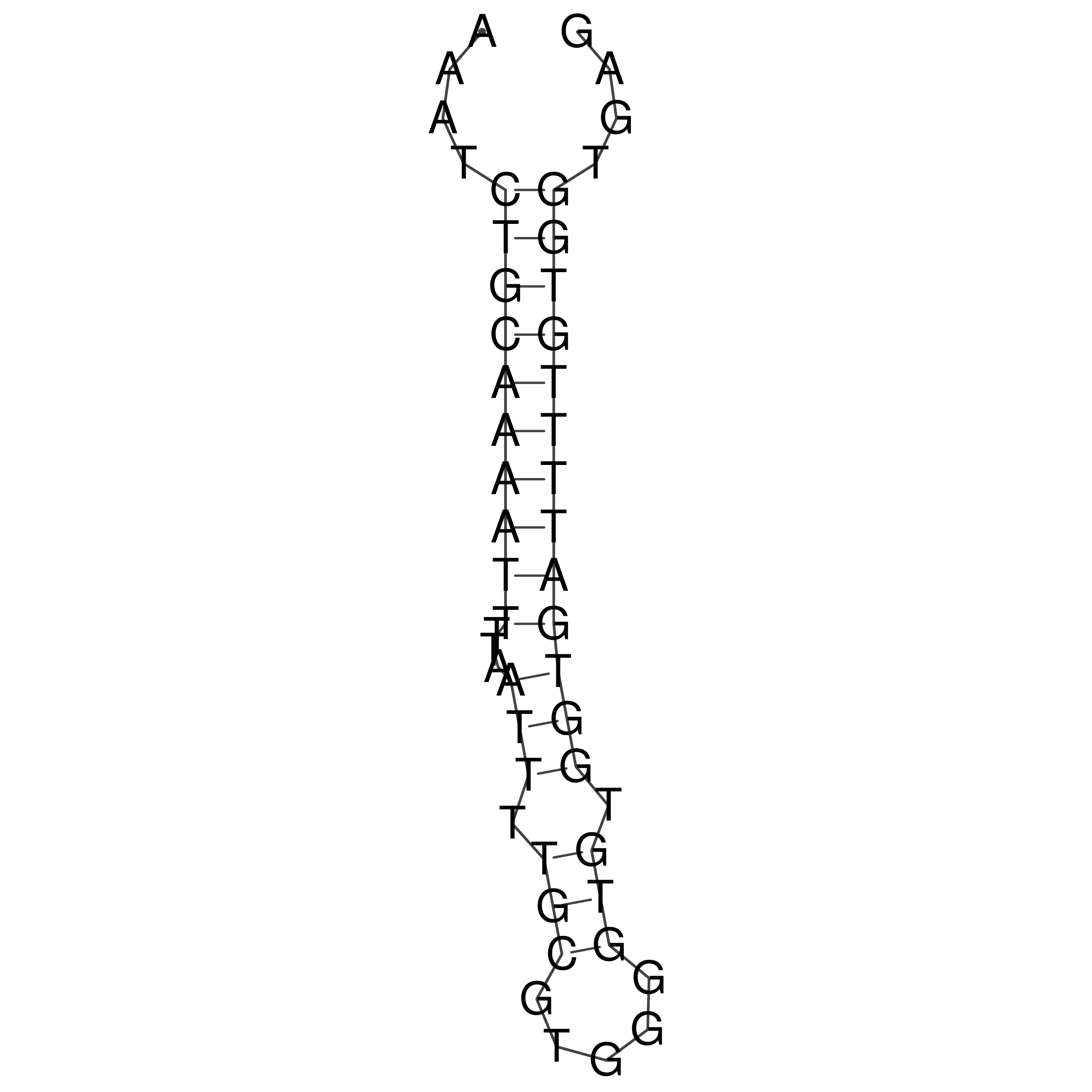
Relaxase
| ID | 3790 | GenBank | WP_240289378 |
| Name | traI_MKZ43_RS27830_bat15|unnamed2 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190786.04 Da Isoelectric Point: 6.4844
>WP_240289378.1 conjugative transfer relaxase/helicase TraI [Klebsiella pneumoniae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGMFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGASGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQKSVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNAPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKKAEQTAREMAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGMFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRHNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGASGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQKSVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTSFSVVSE
QLKTRSGETDLDAAIAQQKAGLRTPAEQAIHLAIPLLESEKLTFSRPQLLATALETGGGKVPMADIDTTI
QTQIRSGQLLNAPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKKAEQTAREMAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3784 | GenBank | WP_032495268 |
| Name | traD_MKZ43_RS27835_bat15|unnamed2 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85986.98 Da Isoelectric Point: 5.0632
>WP_032495268.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRRLDTRVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIPRRLDTRVDARLSALLEAREAEGSLARALFTPDTPEPGPADTDSHASEQPEPVSPPA
PAVMTVTPAPVKSPPTTKRPAAEPSVRATEPPVLRGTTVPLIKPKAAAAATAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 66117..95114
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MKZ43_RS27835 (MKZ43_27835) | 66117..68429 | - | 2313 | WP_032495268 | type IV conjugative transfer system coupling protein TraD | virb4 |
| MKZ43_RS27840 (MKZ43_27840) | 68558..69247 | - | 690 | WP_004198206 | hypothetical protein | - |
| MKZ43_RS27845 (MKZ43_27845) | 69439..70170 | - | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| MKZ43_RS27850 (MKZ43_27850) | 70354..70902 | - | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| MKZ43_RS27855 (MKZ43_27855) | 70915..73764 | - | 2850 | WP_004152624 | conjugal transfer mating-pair stabilization protein TraG | traG |
| MKZ43_RS27860 (MKZ43_27860) | 73764..75143 | - | 1380 | WP_004165169 | conjugal transfer pilus assembly protein TraH | traH |
| MKZ43_RS27865 (MKZ43_27865) | 75121..75564 | - | 444 | WP_004152679 | F-type conjugal transfer protein TrbF | - |
| MKZ43_RS27870 (MKZ43_27870) | 75610..76167 | - | 558 | WP_004152678 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| MKZ43_RS27875 (MKZ43_27875) | 76139..76378 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| MKZ43_RS27880 (MKZ43_27880) | 76389..77141 | - | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| MKZ43_RS27885 (MKZ43_27885) | 77162..77488 | - | 327 | WP_004152676 | hypothetical protein | - |
| MKZ43_RS27890 (MKZ43_27890) | 77501..77749 | - | 249 | WP_004152675 | hypothetical protein | - |
| MKZ43_RS27895 (MKZ43_27895) | 77727..77981 | - | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| MKZ43_RS27900 (MKZ43_27900) | 78013..79968 | - | 1956 | WP_004152673 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| MKZ43_RS27905 (MKZ43_27905) | 80027..80665 | - | 639 | WP_004152672 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| MKZ43_RS27910 (MKZ43_27910) | 80678..81667 | - | 990 | WP_011977785 | conjugal transfer pilus assembly protein TraU | traU |
| MKZ43_RS27915 (MKZ43_27915) | 81664..82053 | - | 390 | WP_004152508 | hypothetical protein | - |
| MKZ43_RS27920 (MKZ43_27920) | 82095..82721 | - | 627 | WP_004152507 | type-F conjugative transfer system protein TraW | traW |
| MKZ43_RS27925 (MKZ43_27925) | 82721..83110 | - | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| MKZ43_RS27930 (MKZ43_27930) | 83110..85749 | - | 2640 | WP_004152505 | type IV secretion system protein TraC | virb4 |
| MKZ43_RS27935 (MKZ43_27935) | 85821..86219 | - | 399 | WP_011977783 | hypothetical protein | - |
| MKZ43_RS27940 (MKZ43_27940) | 86227..86616 | - | 390 | WP_004153076 | hypothetical protein | - |
| MKZ43_RS27945 (MKZ43_27945) | 86659..87063 | - | 405 | WP_004152503 | hypothetical protein | - |
| MKZ43_RS27950 (MKZ43_27950) | 87130..87441 | - | 312 | WP_004152502 | hypothetical protein | - |
| MKZ43_RS27955 (MKZ43_27955) | 87442..87660 | - | 219 | WP_004152501 | hypothetical protein | - |
| MKZ43_RS27960 (MKZ43_27960) | 87766..88176 | - | 411 | WP_004152499 | hypothetical protein | - |
| MKZ43_RS27965 (MKZ43_27965) | 88308..88892 | - | 585 | WP_004161368 | type IV conjugative transfer system lipoprotein TraV | traV |
| MKZ43_RS27970 (MKZ43_27970) | 89006..90430 | - | 1425 | WP_004155033 | F-type conjugal transfer pilus assembly protein TraB | traB |
| MKZ43_RS27975 (MKZ43_27975) | 90430..91170 | - | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| MKZ43_RS27980 (MKZ43_27980) | 91157..91723 | - | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| MKZ43_RS27985 (MKZ43_27985) | 91743..92048 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| MKZ43_RS27990 (MKZ43_27990) | 92062..92430 | - | 369 | WP_004152496 | type IV conjugative transfer system pilin TraA | - |
| MKZ43_RS27995 (MKZ43_27995) | 92484..92870 | - | 387 | WP_004152495 | TraY domain-containing protein | - |
| MKZ43_RS28000 (MKZ43_28000) | 92949..93635 | - | 687 | WP_004152494 | transcriptional regulator TraJ family protein | - |
| MKZ43_RS28005 (MKZ43_28005) | 93809..94207 | - | 399 | WP_004152493 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| MKZ43_RS28010 (MKZ43_28010) | 94629..95114 | + | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| MKZ43_RS28015 (MKZ43_28015) | 95147..95476 | - | 330 | WP_011977736 | DUF5983 family protein | - |
| MKZ43_RS28020 (MKZ43_28020) | 95509..96330 | - | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| MKZ43_RS28025 (MKZ43_28025) | 97150..97983 | - | 834 | WP_004152751 | N-6 DNA methylase | - |
| MKZ43_RS28030 (MKZ43_28030) | 98034..98180 | - | 147 | WP_004152750 | hypothetical protein | - |
| MKZ43_RS28035 (MKZ43_28035) | 98275..98622 | - | 348 | WP_004152749 | hypothetical protein | - |
| MKZ43_RS28040 (MKZ43_28040) | 98679..99032 | - | 354 | WP_004152748 | hypothetical protein | - |
| MKZ43_RS28045 (MKZ43_28045) | 99662..100012 | - | 351 | WP_004153414 | hypothetical protein | - |
Host bacterium
| ID | 6005 | GenBank | NZ_CP092767 |
| Plasmid name | bat15|unnamed2 | Incompatibility group | IncFIB |
| Plasmid size | 118949 bp | Coordinate of oriT [Strand] | 94507..94556 [+] |
| Host baterium | Klebsiella pneumoniae strain bat15 |
Cargo genes
| Drug resistance gene | blaTEM-1A, blaOXA-9, blaKPC-3 |
| Virulence gene | - |
| Metal resistance gene | merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |