Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105537 |
| Name | oriT_pSymA |
| Organism | Sinorhizobium meliloti 2011 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_020527 (515299..515327 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file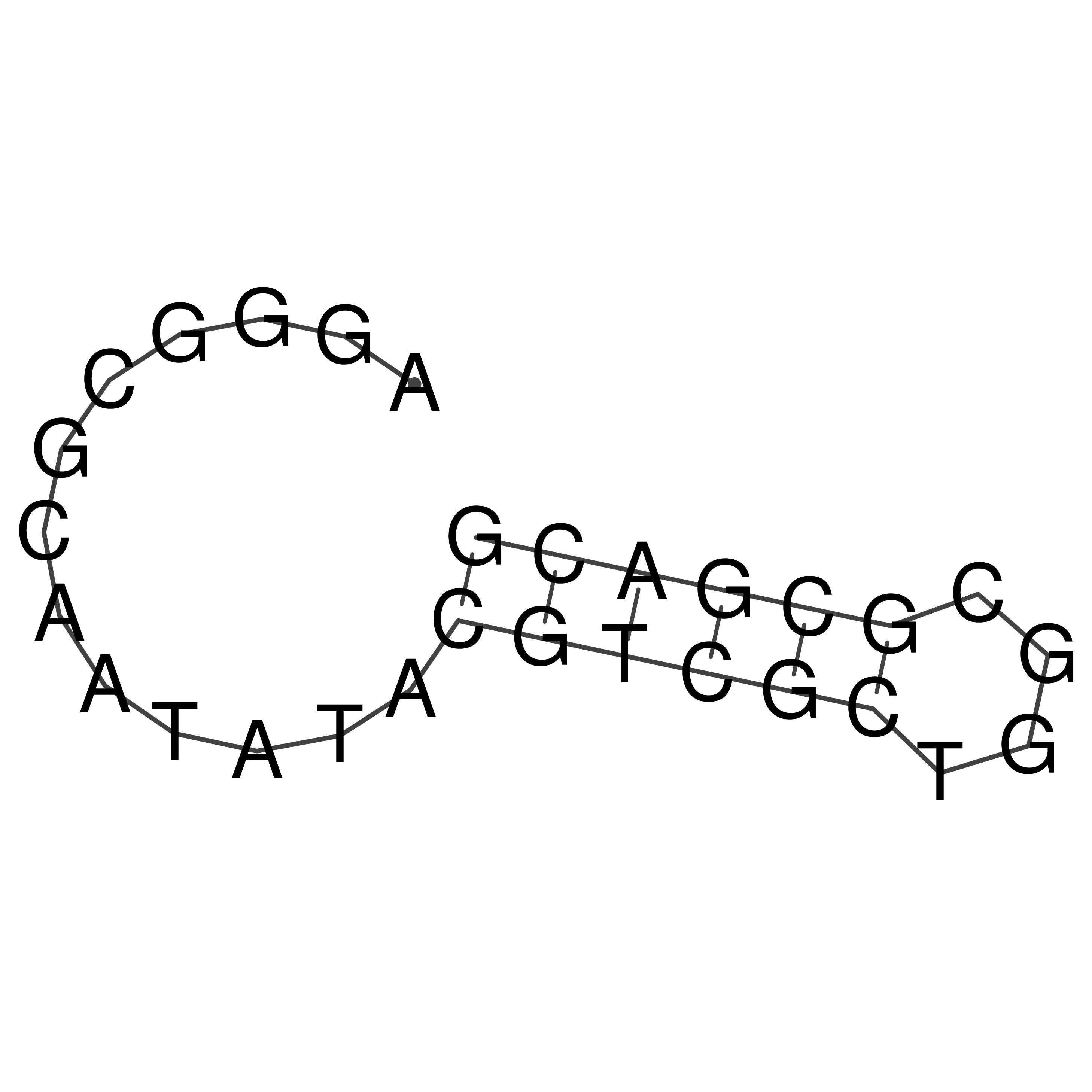
Relaxase
| ID | 3776 | GenBank | WP_015445430 |
| Name | traA_SM2011_RS02575_pSymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169927.57 Da Isoelectric Point: 9.4213
>WP_015445430.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLRAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVITPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDLEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTIVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALAALNALASDAAIEPRKLAEDLGKAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRGITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDQRFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLRAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVITPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDLEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTIVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALAALNALASDAAIEPRKLAEDLGKAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRGITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDQRFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3771 | GenBank | WP_010967483 |
| Name | traG_SM2011_RS02560_pSymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70304.29 Da Isoelectric Point: 9.5501
>WP_010967483.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQERSASTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQERSASTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 710905..720484
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SM2011_RS03535 (SM2011_a1296) | 706522..707544 | - | 1023 | WP_010967682 | alcohol dehydrogenase AdhP | - |
| SM2011_RS03540 (SM2011_a1297) | 707792..708469 | - | 678 | WP_010967683 | YoaK family protein | - |
| SM2011_RS03545 (SM2011_a1299) | 708721..709332 | + | 612 | WP_010967684 | TetR/AcrR family transcriptional regulator | - |
| SM2011_RS03550 (SM2011_a1301) | 709367..710593 | + | 1227 | WP_010967685 | MFS transporter | - |
| SM2011_RS03555 (SM2011_a1302) | 710905..711894 | - | 990 | WP_010967686 | P-type DNA transfer ATPase VirB11 | virB11 |
| SM2011_RS03560 (SM2011_a1303) | 711905..713077 | - | 1173 | WP_010967687 | type IV secretion system protein VirB10 | virB10 |
| SM2011_RS03565 (SM2011_a1306) | 713087..713941 | - | 855 | WP_010967688 | P-type conjugative transfer protein VirB9 | virB9 |
| SM2011_RS03570 (SM2011_a1308) | 713941..714612 | - | 672 | WP_010967689 | virB8 family protein | virB8 |
| SM2011_RS03575 (SM2011_a1310) | 714617..714931 | - | 315 | WP_010967690 | hypothetical protein | - |
| SM2011_RS03580 (SM2011_a1311) | 714928..715863 | - | 936 | WP_013845459 | type IV secretion system protein | virB6 |
| SM2011_RS03585 | 715869..716102 | - | 234 | WP_013845460 | EexN family lipoprotein | - |
| SM2011_RS03590 (SM2011_a1313) | 716099..716800 | - | 702 | WP_010967692 | P-type DNA transfer protein VirB5 | virB5 |
| SM2011_RS03595 (SM2011_a1315) | 716800..719196 | - | 2397 | WP_014528660 | VirB4 family type IV secretion system protein | virb4 |
| SM2011_RS03600 (SM2011_a1318) | 719171..719512 | - | 342 | WP_010967694 | type IV secretion system protein VirB3 | virB3 |
| SM2011_RS03605 (SM2011_a1319) | 719517..719816 | - | 300 | WP_010967695 | TrbC/VirB2 family protein | virB2 |
| SM2011_RS03610 (SM2011_a1321) | 719813..720484 | - | 672 | WP_010967696 | lytic transglycosylase domain-containing protein | virB1 |
| SM2011_RS32075 (SM2011_a1322) | 720488..721006 | - | 519 | WP_010967697 | hypothetical protein | - |
| SM2011_RS03615 (SM2011_a1323) | 721103..721471 | + | 369 | WP_010967698 | transcriptional regulator | - |
| SM2011_RS03620 (SM2011_a1325) | 721732..721983 | - | 252 | WP_010967699 | hypothetical protein | - |
| SM2011_RS03625 (SM2011_a1326) | 722057..722704 | - | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| SM2011_RS03630 (SM2011_a1327) | 722754..723602 | - | 849 | WP_010967701 | alpha/beta fold hydrolase | - |
Host bacterium
| ID | 5975 | GenBank | NC_020527 |
| Plasmid name | pSymA | Incompatibility group | - |
| Plasmid size | 1352561 bp | Coordinate of oriT [Strand] | 515299..515327 [+] |
| Host baterium | Sinorhizobium meliloti 2011 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP, nia, ncrA |
| Degradation gene | - |
| Symbiosis gene | nodD, fixN, fixO, fixU, nifB, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE, nifX, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, nifN, nodM, fixS, fixG |
| Anti-CRISPR | AcrIIA9 |