Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105504 |
| Name | oriT_pL21581-3 |
| Organism | Carnobacterium maltaromaticum strain TMW 2.1581 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP016847 (39439..39575 [-], 137 nt) |
| oriT length | 137 nt |
| IRs (inverted repeats) | 81..86, 88..93 (TTTGTA..TACAAA) 21..28, 42..49 (AAAAGGGG..CCCCTTTT) 25..30, 39..44 (GGGGAA..TTCCCC) |
| Location of nic site | 104..105 |
| Conserved sequence flanking the nic site |
GCTTGCAGTA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 137 nt
>oriT_pL21581-3
CGACACAATCCAAAGGGGATAAAAGGGGAAAGTGAAACTTCCCCCTTTTCAAGCAACTTTGTAATACAAGAACGAAGTCATTTGTATTACAAAGTGATAGCTTGCAGTATTTATGGCTTTATATGGTCTATTTTGTT
CGACACAATCCAAAGGGGATAAAAGGGGAAAGTGAAACTTCCCCCTTTTCAAGCAACTTTGTAATACAAGAACGAAGTCATTTGTATTACAAAGTGATAGCTTGCAGTATTTATGGCTTTATATGGTCTATTTTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file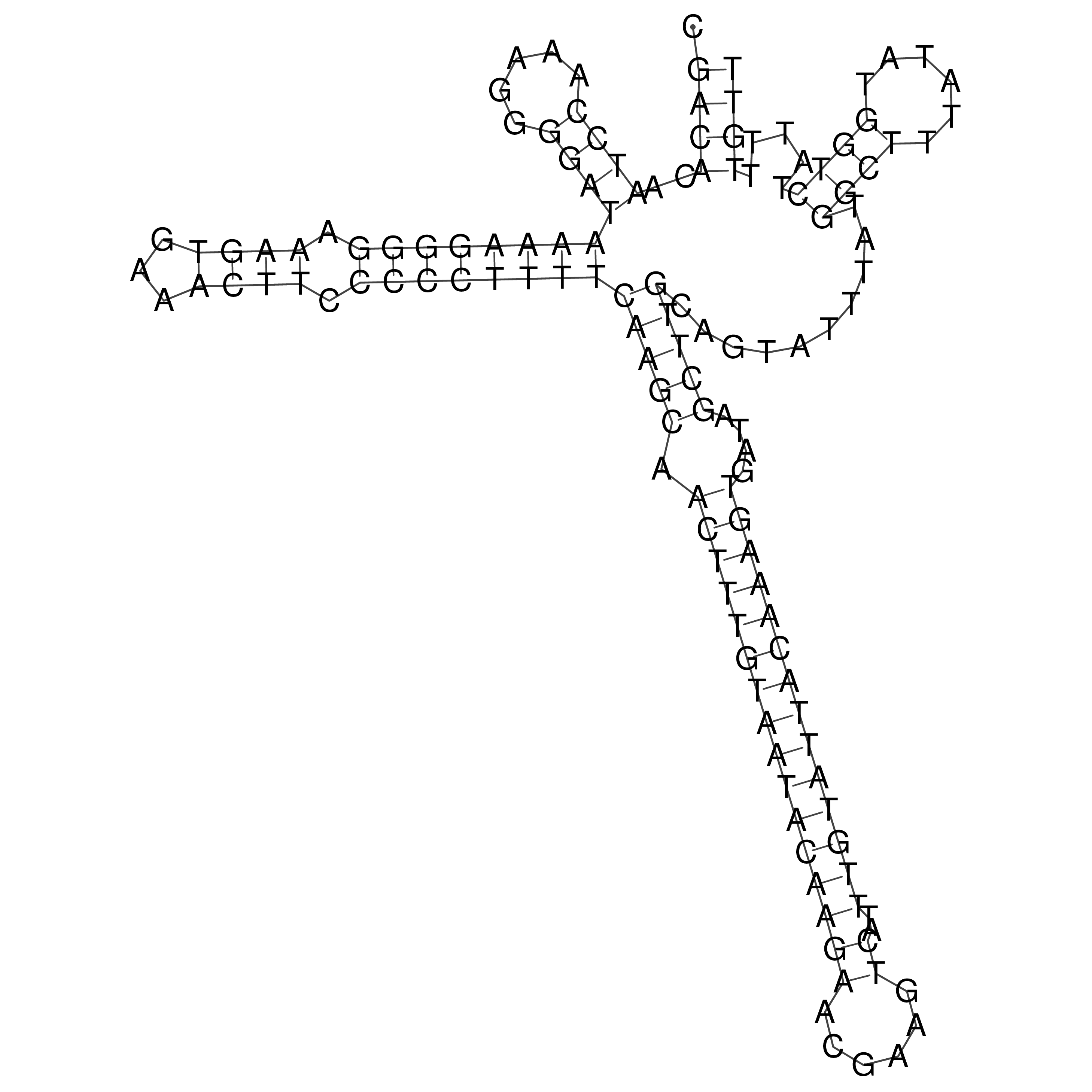
Relaxase
| ID | 3747 | GenBank | WP_035063709 |
| Name | Replic_Relax_BFC23_RS16930_pL21581-3 |
UniProt ID | _ |
| Length | 267 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 267 a.a. Molecular weight: 32077.08 Da Isoelectric Point: 8.2273
>WP_035063709.1 MULTISPECIES: replication-relaxation family protein [Carnobacterium]
MEALLANYYVEFKPRVNTDWIILCILHELRIVSFKQIYELINLTFPIAEKNLRRQINQLKDKELIDFITD
DKLGNAHYYFMTRDGHNSIGGYYPYPKVPQYNLFHHLMVTNALINVLTLFNEHPHLLYVQSERRQVFEVK
DCDPSKKGKIYNVSDFLFCFEAKSELKINWYFEIELTMKTKNRYTNNIFPKYLALLQRDKNARLIYVTPS
KSIFRELELFKGFYLHKKKKEVGEKADELFERLHIFSSESFEDNLKKIIAEDEFINW
MEALLANYYVEFKPRVNTDWIILCILHELRIVSFKQIYELINLTFPIAEKNLRRQINQLKDKELIDFITD
DKLGNAHYYFMTRDGHNSIGGYYPYPKVPQYNLFHHLMVTNALINVLTLFNEHPHLLYVQSERRQVFEVK
DCDPSKKGKIYNVSDFLFCFEAKSELKINWYFEIELTMKTKNRYTNNIFPKYLALLQRDKNARLIYVTPS
KSIFRELELFKGFYLHKKKKEVGEKADELFERLHIFSSESFEDNLKKIIAEDEFINW
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 3739 | GenBank | WP_051926640 |
| Name | t4cp2_BFC23_RS16925_pL21581-3 |
UniProt ID | _ |
| Length | 714 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 714 a.a. Molecular weight: 81356.94 Da Isoelectric Point: 5.2483
>WP_051926640.1 TraM recognition domain-containing protein [Carnobacterium maltaromaticum]
MNDDDNLYNGLFGNKKNQKNGNENDKTSRYNVISIVVSISFVLLYPFAIFGIFPVLYFIYLDKQQRLENV
HDIEFKSFLQKGNSTFGLISGGLILVNILSSIYLIPRGYLSAYLLFPLNMIHHALVFSWQTVVALLLGGA
GMASIMLTIFSFIEGRKVVSKKSQIAKVKESKAYKKRLSNKFEESKNFVEEYEEAYATAKLEGNVTKLDE
LAHNILLGTDEYGKPYFITLKELNQHALLPATTGGGKTVVLQIIVEHCAKFNSPCLIIDGKGAKDTRLAV
SDIAGKYNREVKEFTDSGDISYNPLKYGNSIEVRDKLVSLAETESIFYSGANKQLLMGTTQLLDVFNIER
DLENMSDYFSPRKVLLLFINELLLIYPDLLVFEVEESPKNKSKKSVKKEVEVVEKAEVKDSASSKYDQIE
DDGVATEVVETITANPETMKLEDMYWVISNKKKRLKKSSLRMFEQLFARYEHKDNPFYLYATAESLQTNF
YMLLDSELSHLFDTKTNKNELDLMEATRKNEIIYVALDGLIYTEYIHTLAQFMIADINHLASERYEITED
FPFVVLFDEPSAYLSDKFIDTVNKTRGAGVHALFAPQTLADIEAIDPILLKQLIGNTNTYIVGQTNEKSE
IDYWSELFGSYDDIEITSVTQQEAGFSDVNKTDWTGEKGTKRDVENFIVHPNNIKELRTGEFYILRKGNN
THEPIRKVYMRKPI
MNDDDNLYNGLFGNKKNQKNGNENDKTSRYNVISIVVSISFVLLYPFAIFGIFPVLYFIYLDKQQRLENV
HDIEFKSFLQKGNSTFGLISGGLILVNILSSIYLIPRGYLSAYLLFPLNMIHHALVFSWQTVVALLLGGA
GMASIMLTIFSFIEGRKVVSKKSQIAKVKESKAYKKRLSNKFEESKNFVEEYEEAYATAKLEGNVTKLDE
LAHNILLGTDEYGKPYFITLKELNQHALLPATTGGGKTVVLQIIVEHCAKFNSPCLIIDGKGAKDTRLAV
SDIAGKYNREVKEFTDSGDISYNPLKYGNSIEVRDKLVSLAETESIFYSGANKQLLMGTTQLLDVFNIER
DLENMSDYFSPRKVLLLFINELLLIYPDLLVFEVEESPKNKSKKSVKKEVEVVEKAEVKDSASSKYDQIE
DDGVATEVVETITANPETMKLEDMYWVISNKKKRLKKSSLRMFEQLFARYEHKDNPFYLYATAESLQTNF
YMLLDSELSHLFDTKTNKNELDLMEATRKNEIIYVALDGLIYTEYIHTLAQFMIADINHLASERYEITED
FPFVVLFDEPSAYLSDKFIDTVNKTRGAGVHALFAPQTLADIEAIDPILLKQLIGNTNTYIVGQTNEKSE
IDYWSELFGSYDDIEITSVTQQEAGFSDVNKTDWTGEKGTKRDVENFIVHPNNIKELRTGEFYILRKGNN
THEPIRKVYMRKPI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 5942 | GenBank | NZ_CP016847 |
| Plasmid name | pL21581-3 | Incompatibility group | - |
| Plasmid size | 69778 bp | Coordinate of oriT [Strand] | 39439..39575 [-] |
| Host baterium | Carnobacterium maltaromaticum strain TMW 2.1581 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |