Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105496 |
| Name | oriT_pKAM644_4 |
| Organism | Klebsiella quasipneumoniae subsp. quasipneumoniae strain KAM644 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_AP026411 (44270..44318 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pKAM644_4
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file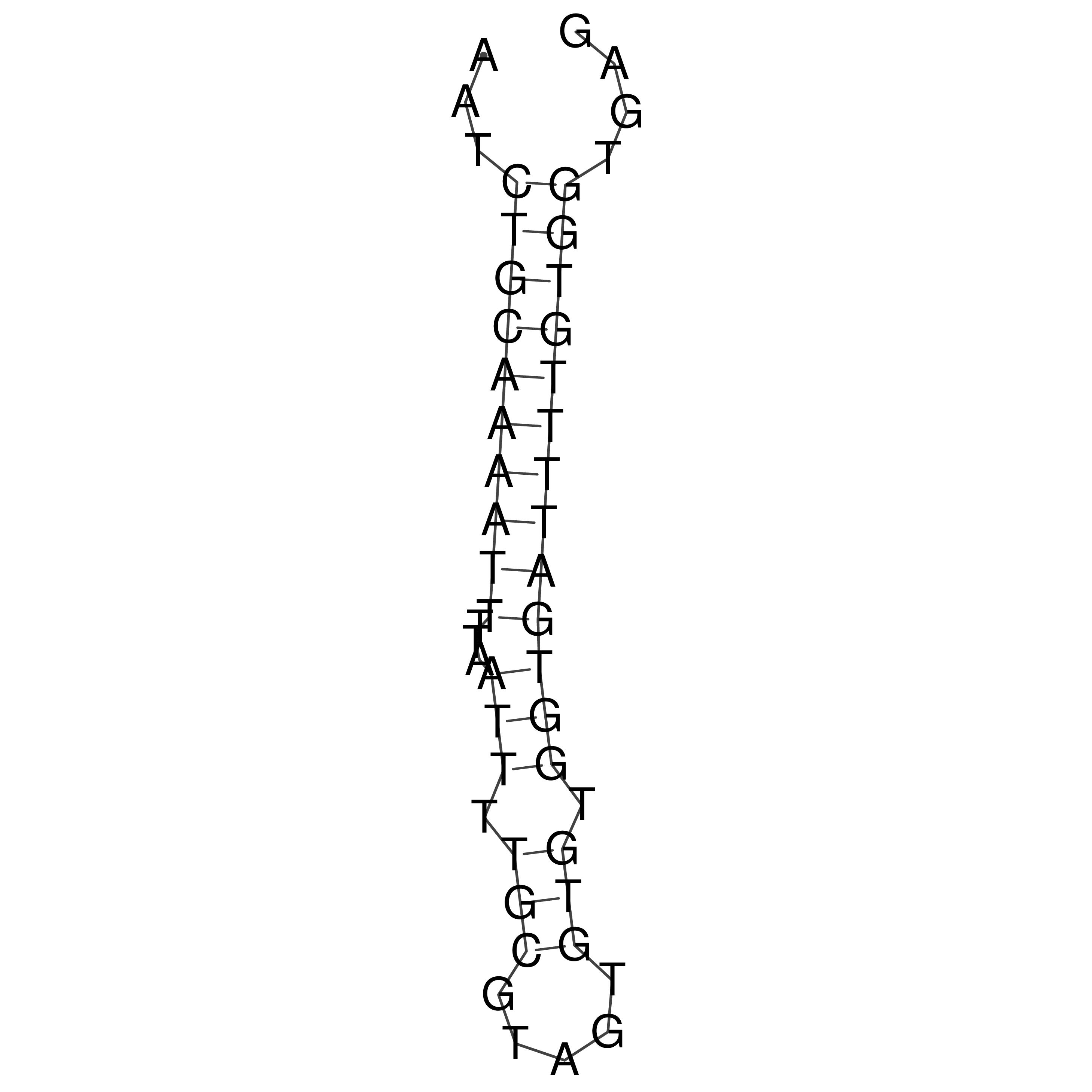
Relaxase
| ID | 3743 | GenBank | WP_276097507 |
| Name | traI_P4I75_RS29725_pKAM644_4 |
UniProt ID | _ |
| Length | 1744 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1744 a.a. Molecular weight: 189524.89 Da Isoelectric Point: 5.9131
>WP_276097507.1 conjugative transfer relaxase/helicase TraI [Klebsiella quasipneumoniae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESDGVSETVLTGNLIIARFNHDT
SRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVESLGYRTVDA
GKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNTLKETGFDI
RGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEAKDGVFELA
RKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPFSDAVSVLA
QDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTGKSALQDGT
AFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYRWQGGHQTT
ADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDTTITALTPV
WLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVDSQWTLFRA
DTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIKVGHGWVES
PGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQLKSRSGET
DLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQAQIRSGQL
LNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATRMILESTDRF
TVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHDTQLLQRNG
QTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRSAADVAIMK
EIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKAIEKALSEGKTL
PEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLPVLVTSNIR
DGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLYRQEKITVS
QGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITAHGAQGASE
PYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEPRNDRAVKT
ADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIWLSPLTDRD
GRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRPWNPGAMTG
GRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPADRVKEVIG
DVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLGGD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAEALGLEGKVDKQIFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESDGVSETVLTGNLIIARFNHDT
SRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIAFGKIYQNTLRADVESLGYRTVDA
GKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNTLKETGFDI
RGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEAKDGVFELA
RKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTSDASVVRQAPFSDAVSVLA
QDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTGKSALQDGT
AFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYRWQGGHQTT
ADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDTTITALTPV
WLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVDSQWTLFRA
DTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIKVGHGWVES
PGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQLKSRSGET
DLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQAQIRSGQL
LNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATRMILESTDRF
TVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHDTQLLQRNG
QTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRSAADVAIMK
EIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKAIEKALSEGKTL
PEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLPVLVTSNIR
DGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLYRQEKITVS
QGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITAHGAQGASE
PYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEPRNDRAVKT
ADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIWLSPLTDRD
GRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRPWNPGAMTG
GRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPADRVKEVIG
DVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1693 | GenBank | WP_004197733 |
| Name | WP_004197733_pKAM644_4 |
UniProt ID | A0A0E3KB00 |
| Length | 129 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 129 a.a. Molecular weight: 14835.90 Da Isoelectric Point: 4.6012
>WP_004197733.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Bacteria]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKILGIECLSPHVKDDPRWQWKSMVLNIQEDVQEAMRNFFPDEDAEGE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A0E3KB00 |
T4CP
| ID | 3737 | GenBank | WP_009309880 |
| Name | traD_P4I75_RS29720_pKAM644_4 |
UniProt ID | _ |
| Length | 769 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 769 a.a. Molecular weight: 85571.58 Da Isoelectric Point: 5.1733
>WP_009309880.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPDDAGSHAGEQPEPVSQPA
PAVMTVTPAPVKSPPTTKRPAAEPSARTAEPPVLRVTTVPLIKPKAAAAAGTAATASSAGTPAAAAGGTE
QVLAQQSAEQGQAMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDLEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 46732..72734
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| P4I75_RS29540 (KAM644c_57440) | 42055..42285 | + | 231 | WP_020314756 | hypothetical protein | - |
| P4I75_RS29545 | 42370..42489 | + | 120 | Protein_56 | SAM-dependent DNA methyltransferase | - |
| P4I75_RS29550 (KAM644c_57450) | 43138..43680 | + | 543 | WP_020314754 | antirestriction protein | - |
| P4I75_RS29555 (KAM644c_57460) | 43703..44125 | - | 423 | WP_020315406 | transglycosylase SLT domain-containing protein | - |
| P4I75_RS29560 (KAM644c_57470) | 44630..45019 | + | 390 | WP_004197733 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| P4I75_RS29565 (KAM644c_57480) | 45258..45932 | + | 675 | WP_004197737 | hypothetical protein | - |
| P4I75_RS29570 (KAM644c_57490) | 46124..46288 | + | 165 | WP_032444506 | TraY domain-containing protein | - |
| P4I75_RS29575 (KAM644c_57500) | 46350..46718 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| P4I75_RS29580 (KAM644c_57510) | 46732..47037 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| P4I75_RS29585 | 47057..47617 | + | 561 | Protein_64 | type IV conjugative transfer system protein TraE | - |
| P4I75_RS29590 | 47666..48349 | + | 684 | WP_276097508 | type-F conjugative transfer system secretin TraK | traK |
| P4I75_RS29595 (KAM644c_57530) | 48349..49773 | + | 1425 | WP_004194260 | F-type conjugal transfer pilus assembly protein TraB | traB |
| P4I75_RS29600 | 49887..50471 | + | 585 | WP_009309868 | type IV conjugative transfer system lipoprotein TraV | traV |
| P4I75_RS29605 (KAM644c_57540) | 50603..51013 | + | 411 | WP_009309869 | hypothetical protein | - |
| P4I75_RS29610 (KAM644c_57550) | 51078..52112 | - | 1035 | WP_116994126 | IS630 family transposase | - |
| P4I75_RS29615 (KAM644c_57560) | 52221..52439 | + | 219 | WP_004195468 | hypothetical protein | - |
| P4I75_RS29620 (KAM644c_57570) | 52440..52751 | + | 312 | WP_004195465 | hypothetical protein | - |
| P4I75_RS29625 (KAM644c_57580) | 52818..53222 | + | 405 | WP_004197817 | hypothetical protein | - |
| P4I75_RS29630 | 53599..53997 | + | 399 | WP_023179972 | hypothetical protein | - |
| P4I75_RS29635 (KAM644c_57590) | 54069..56708 | + | 2640 | WP_004197813 | type IV secretion system protein TraC | virb4 |
| P4I75_RS29640 (KAM644c_57600) | 56708..57097 | + | 390 | WP_004197815 | type-F conjugative transfer system protein TrbI | - |
| P4I75_RS29645 (KAM644c_57610) | 57097..57723 | + | 627 | WP_009309871 | type-F conjugative transfer system protein TraW | traW |
| P4I75_RS29650 (KAM644c_57620) | 57765..58154 | + | 390 | WP_004194992 | hypothetical protein | - |
| P4I75_RS29655 (KAM644c_57630) | 58151..59140 | + | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| P4I75_RS29660 (KAM644c_57640) | 59153..59791 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| P4I75_RS29665 (KAM644c_57650) | 59850..61805 | + | 1956 | WP_276097503 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| P4I75_RS29670 (KAM644c_57660) | 61837..62091 | + | 255 | WP_004195500 | conjugal transfer protein TrbE | - |
| P4I75_RS29675 (KAM644c_57670) | 62069..62317 | + | 249 | WP_004152675 | hypothetical protein | - |
| P4I75_RS29680 (KAM644c_57680) | 62330..62656 | + | 327 | WP_004194451 | hypothetical protein | - |
| P4I75_RS29685 (KAM644c_57690) | 62677..63429 | + | 753 | WP_009309874 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| P4I75_RS29690 (KAM644c_57700) | 63440..63679 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| P4I75_RS29695 (KAM644c_57710) | 63651..64223 | + | 573 | WP_009309876 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| P4I75_RS29700 (KAM644c_57720) | 64216..64644 | + | 429 | WP_004152689 | hypothetical protein | - |
| P4I75_RS29705 (KAM644c_57730) | 64631..66001 | + | 1371 | WP_117249438 | conjugal transfer pilus assembly protein TraH | traH |
| P4I75_RS29710 (KAM644c_57740) | 66001..68874 | + | 2874 | WP_276097505 | conjugal transfer mating-pair stabilization protein TraG | traG |
| P4I75_RS29715 (KAM644c_57750) | 69609..70298 | + | 690 | WP_276086049 | hypothetical protein | - |
| P4I75_RS29720 (KAM644c_57760) | 70425..72734 | + | 2310 | WP_009309880 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 5934 | GenBank | NZ_AP026411 |
| Plasmid name | pKAM644_4 | Incompatibility group | IncFII |
| Plasmid size | 82914 bp | Coordinate of oriT [Strand] | 44270..44318 [-] |
| Host baterium | Klebsiella quasipneumoniae subsp. quasipneumoniae strain KAM644 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |