Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105477 |
| Name | oriT_pM084986_1 |
| Organism | Staphylococcus aureus strain RIVM_M084986 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP096529 (33455..33490 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACG..CGTTCGC) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pM084986_1
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
CACGCGAACGGAACGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file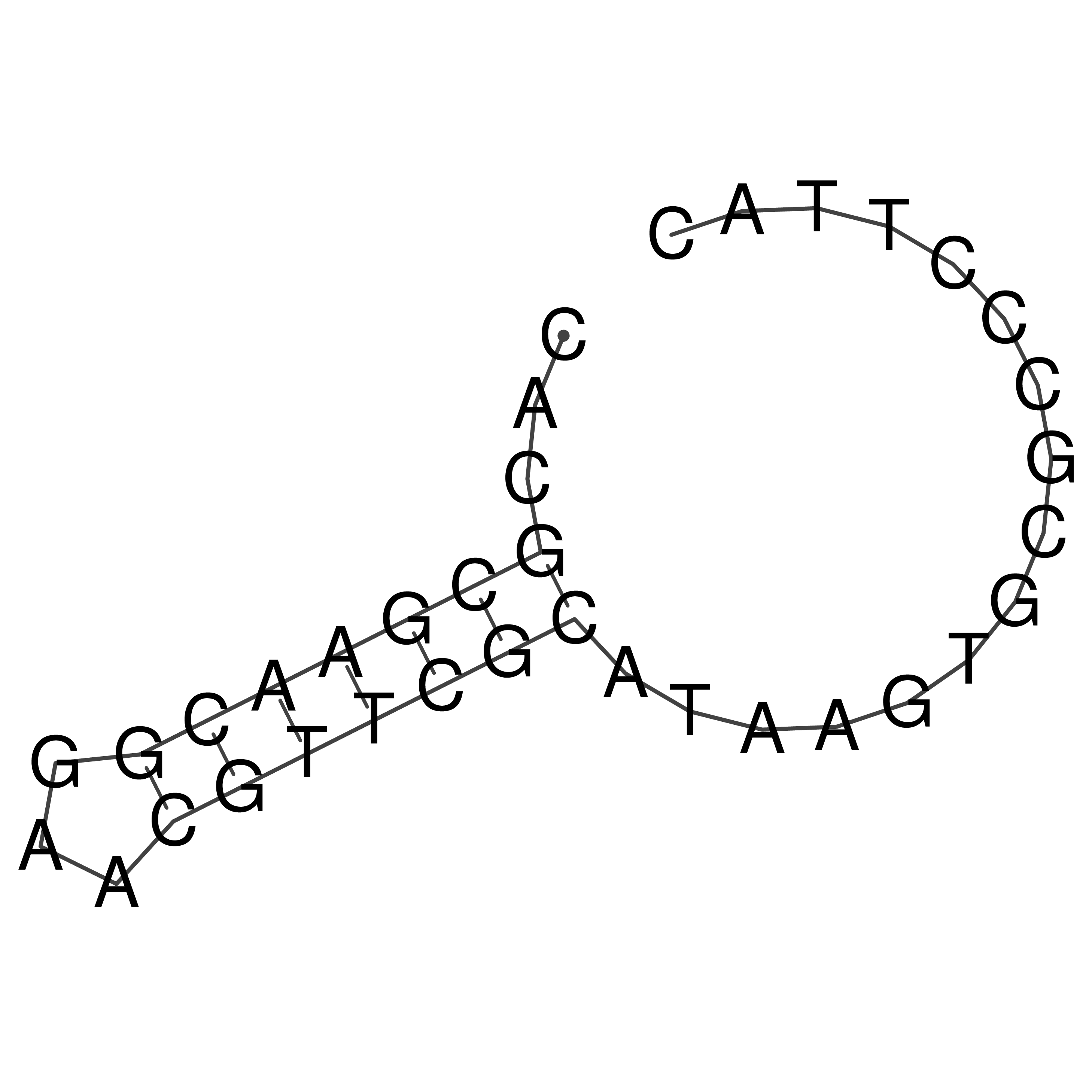
Relaxase
| ID | 3732 | GenBank | WP_231493169 |
| Name | MobA_MobL_M1F50_RS13980_pM084986_1 |
UniProt ID | _ |
| Length | 665 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 79066.24 Da Isoelectric Point: 6.0866
>WP_231493169.1 MULTISPECIES: MobA/MobL family protein [Staphylococcaceae]
MAMYHFQNKFVSKAKGQSATAKAAYNSASKIKDYKENKFKDYSNKQCDYSEIILPENADNKFKDREYLWN
KVNDVENRKNSQVAREIIIGLPNEFDSESNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSSDSYKNQNIDLEPTKKEGW
KARKFEDETGHKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYEENERLYDLKQKTALITDDEDRLNKIENIEDRQRKLESINEVFEKQSKIFFDKNYPDQKLNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTIIGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHDDLQDKLKIDYYTNKLDILRTAENTLEDYYDVQIKELFTNDEDYKAFNGVTDIKEKQ
QLIDFKTYHGTENTLKMLETGDFIPKYSNEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLNEY
DFDYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILIDKETYNNYNRKQQAYGLINAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
MAMYHFQNKFVSKAKGQSATAKAAYNSASKIKDYKENKFKDYSNKQCDYSEIILPENADNKFKDREYLWN
KVNDVENRKNSQVAREIIIGLPNEFDSESNIELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWENVQNKHLEKNGFSVRVSSDSYKNQNIDLEPTKKEGW
KARKFEDETGHKSSISKHNESIKKRNQQKIDKMFNEVDDMKSHKLNAFSYMNKSDSTTLKNMAKDLKIYV
TPINMYEENERLYDLKQKTALITDDEDRLNKIENIEDRQRKLESINEVFEKQSKIFFDKNYPDQKLNYSD
DEKIFITRTILNDRDVLPANHELEDIVKEKRIKEAQISLNTIIGNRDISLESIEKESNFFADKLSNILEK
NNLSFDDVLENKHDDLQDKLKIDYYTNKLDILRTAENTLEDYYDVQIKELFTNDEDYKAFNGVTDIKEKQ
QLIDFKTYHGTENTLKMLETGDFIPKYSNEDRKYITEQVKLLQEKEFKPNKNQHDKFVFGAIQKKLLNEY
DFDYSDNNDLKHLYQESNEVGDEISKDNIEEFYEGNDILIDKETYNNYNRKQQAYGLINAGLDSMIFNFN
EIFRERMPKYINHQYKGKNHSKQRHELKNKRGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3725 | GenBank | WP_231493179 |
| Name | t4cp2_M1F50_RS13890_pM084986_1 |
UniProt ID | _ |
| Length | 548 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 548 a.a. Molecular weight: 62690.58 Da Isoelectric Point: 6.5430
>WP_231493179.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Staphylococcaceae]
MTTLLGELESKATSLNKKNLVIQDFDTKCPNRNIFVVGGPGSYKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKINKDVWYKSSVGLL
NSLLLYAQYEFEPEKRNIGNIIKFIQNNKPNHDEEGSVELDNRFNELPKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ANGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKDELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMKQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETFIAPQNEYKRKKNQSMIDKHNRLKEEWDKKQEKIKQQKLEEYKEEQ
RQKALEEEQQSKEEQQNEEVNESKDTNNEPPEDKKEEKQKINDSKKAFYEKLKQKQAK
MTTLLGELESKATSLNKKNLVIQDFDTKCPNRNIFVVGGPGSYKSAGYVIPNVIVKNQQSIVVTDPSGEV
YEKTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDCSIVAELIIKSAGDSKINKDVWYKSSVGLL
NSLLLYAQYEFEPEKRNIGNIIKFIQNNKPNHDEEGSVELDNRFNELPKDHPARESYEFGFAVSEGRTRA
SIILTFVADLRNYIDNEIRSYTSDSDFDLRDVGLRKTIIYVMLPVLGNTVQSLSSLFFSQMFQQLYRLGD
ANGARLPVPVDFLLDEWPNIGAIPDYEETLATCRKYGISITTIVQSISQAVDKYNKDKANAIIGNHAVIL
CLGNVNNDTAKYIKDELGNATVEFETSSEGSSSGKDSRSKSSNKNVSYTGRALLNEDEISNMKQDKAILI
TKNRNPKIIKKLAYFKMFPNLTETFIAPQNEYKRKKNQSMIDKHNRLKEEWDKKQEKIKQQKLEEYKEEQ
RQKALEEEQQSKEEQQNEEVNESKDTNNEPPEDKKEEKQKINDSKKAFYEKLKQKQAK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 16335..27571
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| M1F50_RS13860 (M1F50_13840) | 12360..12539 | - | 180 | WP_013356267 | hypothetical protein | - |
| M1F50_RS13865 (M1F50_13850) | 12783..14156 | - | 1374 | WP_077896374 | ATP-binding protein | - |
| M1F50_RS13870 (M1F50_13855) | 14159..15244 | - | 1086 | WP_000867971 | tyrosine-type recombinase/integrase | - |
| M1F50_RS13875 (M1F50_13860) | 15372..15821 | - | 450 | WP_231493176 | hypothetical protein | - |
| M1F50_RS13880 (M1F50_13865) | 15923..16318 | - | 396 | WP_231493177 | single-stranded DNA-binding protein | - |
| M1F50_RS13885 (M1F50_13870) | 16335..17252 | - | 918 | WP_231493178 | conjugal transfer protein TrbL family protein | trsL |
| M1F50_RS13890 (M1F50_13875) | 17330..18976 | - | 1647 | WP_231493179 | VirD4-like conjugal transfer protein, CD1115 family | - |
| M1F50_RS13895 (M1F50_13880) | 18979..19437 | - | 459 | WP_231493180 | hypothetical protein | trsJ |
| M1F50_RS13900 (M1F50_13885) | 19453..21555 | - | 2103 | WP_231493181 | DNA topoisomerase III | - |
| M1F50_RS13905 (M1F50_13890) | 21571..22056 | - | 486 | WP_231493182 | TrsH/TraH family protein | - |
| M1F50_RS13910 (M1F50_13895) | 22066..23139 | - | 1074 | WP_231493183 | CHAP domain-containing protein | trsG |
| M1F50_RS13915 (M1F50_13900) | 23157..24449 | - | 1293 | WP_231493184 | hypothetical protein | trsF |
| M1F50_RS13920 (M1F50_13905) | 24461..26473 | - | 2013 | WP_231493185 | DUF87 domain-containing protein | virb4 |
| M1F50_RS13925 (M1F50_13910) | 26494..27171 | - | 678 | WP_233670897 | TrsD/TraD family conjugative transfer protein | trsD |
| M1F50_RS13930 (M1F50_13915) | 27164..27571 | - | 408 | WP_231493161 | hypothetical protein | trsC |
| M1F50_RS13935 (M1F50_13920) | 27630..27947 | - | 318 | WP_084756775 | CagC family type IV secretion system protein | - |
| M1F50_RS13940 (M1F50_13925) | 27964..28938 | - | 975 | WP_231493162 | conjugative transfer protein TrsA | - |
| M1F50_RS13945 (M1F50_13930) | 29111..29296 | + | 186 | WP_231493163 | regulator | - |
| M1F50_RS13950 (M1F50_13935) | 29423..30472 | - | 1050 | WP_267440095 | nuclease | - |
| M1F50_RS13955 (M1F50_13940) | 30857..31117 | + | 261 | WP_231493165 | helix-turn-helix transcriptional regulator | - |
| M1F50_RS13960 (M1F50_13945) | 31121..31450 | - | 330 | WP_231493166 | plasmid segregation protein ParR | - |
| M1F50_RS13965 (M1F50_13950) | 31453..32493 | - | 1041 | WP_231493167 | parM protein | - |
Host bacterium
| ID | 5915 | GenBank | NZ_CP096529 |
| Plasmid name | pM084986_1 | Incompatibility group | - |
| Plasmid size | 35612 bp | Coordinate of oriT [Strand] | 33455..33490 [-] |
| Host baterium | Staphylococcus aureus strain RIVM_M084986 |
Cargo genes
| Drug resistance gene | fexA, cfr |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |