Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105437 |
| Name | oriT_pSA-KpST14-3 |
| Organism | Klebsiella pneumoniae subsp. pneumoniae strain SA-KpST14 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP071282 (71654..71703 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pSA-KpST14-3
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTCATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file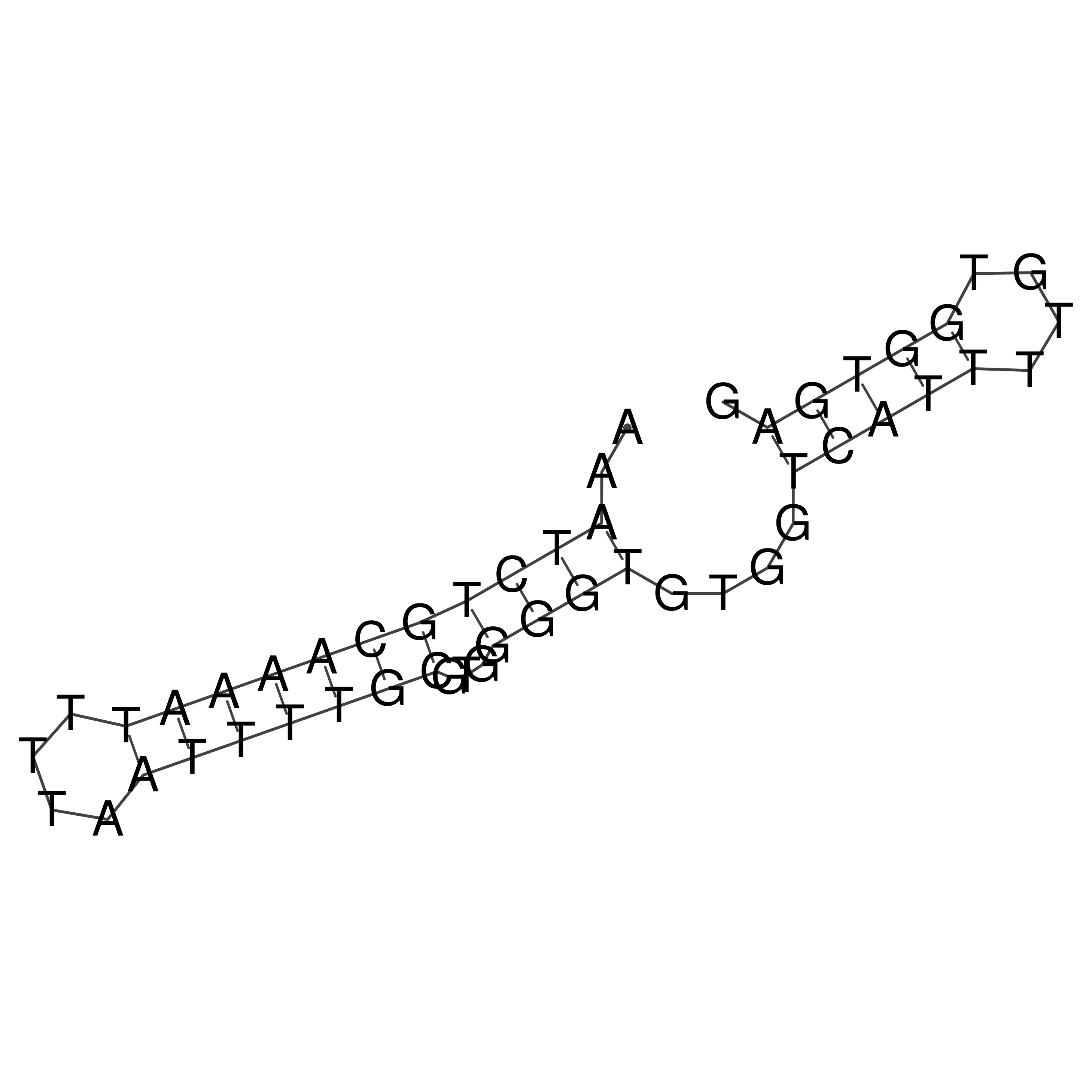
Relaxase
| ID | 3707 | GenBank | WP_043907038 |
| Name | traI_JZ794_RS28295_pSA-KpST14-3 |
UniProt ID | A0A0S1TKY9 |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190458.19 Da Isoelectric Point: 6.0263
>WP_043907038.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSLSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSLSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWLTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPKQEKAIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3694 | GenBank | WP_043907039 |
| Name | traD_JZ794_RS28290_pSA-KpST14-3 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85906.85 Da Isoelectric Point: 5.0577
>WP_043907039.1 type IV conjugative transfer system coupling protein TraD [Klebsiella pneumoniae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 71096..100260
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JZ794_RS28070 (JZ794_28075) | 66113..66463 | + | 351 | WP_004152758 | hypothetical protein | - |
| JZ794_RS28075 (JZ794_28080) | 67099..67455 | + | 357 | WP_004152717 | hypothetical protein | - |
| JZ794_RS28080 (JZ794_28085) | 67516..67728 | + | 213 | WP_004152718 | hypothetical protein | - |
| JZ794_RS28085 (JZ794_28090) | 67739..67963 | + | 225 | WP_004152719 | hypothetical protein | - |
| JZ794_RS28090 (JZ794_28095) | 68044..68364 | + | 321 | WP_004152720 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| JZ794_RS28095 (JZ794_28100) | 68354..68632 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| JZ794_RS28100 (JZ794_28105) | 68633..69046 | + | 414 | WP_004152722 | helix-turn-helix domain-containing protein | - |
| JZ794_RS28105 (JZ794_28110) | 69880..70701 | + | 822 | WP_004178064 | DUF932 domain-containing protein | - |
| JZ794_RS28110 (JZ794_28115) | 70734..71063 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| JZ794_RS28115 (JZ794_28120) | 71096..71581 | - | 486 | WP_004178063 | transglycosylase SLT domain-containing protein | virB1 |
| JZ794_RS28120 (JZ794_28125) | 72015..72407 | + | 393 | WP_004178062 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| JZ794_RS28125 (JZ794_28130) | 72621..73316 | + | 696 | WP_004178061 | transcriptional regulator TraJ family protein | - |
| JZ794_RS28130 (JZ794_28135) | 73400..73771 | + | 372 | WP_004208838 | TraY domain-containing protein | - |
| JZ794_RS28135 (JZ794_28140) | 73825..74193 | + | 369 | WP_004152496 | type IV conjugative transfer system pilin TraA | - |
| JZ794_RS28140 (JZ794_28145) | 74207..74512 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| JZ794_RS28145 (JZ794_28150) | 74532..75098 | + | 567 | WP_004152602 | type IV conjugative transfer system protein TraE | traE |
| JZ794_RS28150 (JZ794_28155) | 75085..75825 | + | 741 | WP_004152601 | type-F conjugative transfer system secretin TraK | traK |
| JZ794_RS28155 (JZ794_28160) | 75825..77249 | + | 1425 | WP_004152600 | F-type conjugal transfer pilus assembly protein TraB | traB |
| JZ794_RS28160 (JZ794_28165) | 77242..77838 | + | 597 | WP_004152599 | conjugal transfer pilus-stabilizing protein TraP | - |
| JZ794_RS28165 (JZ794_28170) | 77861..78430 | + | 570 | WP_004152598 | type IV conjugative transfer system lipoprotein TraV | traV |
| JZ794_RS28170 (JZ794_28175) | 78562..78972 | + | 411 | WP_004152597 | lipase chaperone | - |
| JZ794_RS28175 (JZ794_28180) | 78977..79267 | + | 291 | WP_004152596 | hypothetical protein | - |
| JZ794_RS28180 (JZ794_28185) | 79291..79509 | + | 219 | WP_004171484 | hypothetical protein | - |
| JZ794_RS28185 (JZ794_28190) | 79510..79827 | + | 318 | WP_004152595 | hypothetical protein | - |
| JZ794_RS28190 (JZ794_28195) | 79894..80298 | + | 405 | WP_004152594 | hypothetical protein | - |
| JZ794_RS28195 (JZ794_28200) | 80594..80992 | + | 399 | WP_004153071 | hypothetical protein | - |
| JZ794_RS28200 (JZ794_28205) | 81064..83703 | + | 2640 | WP_004152592 | type IV secretion system protein TraC | virb4 |
| JZ794_RS28205 (JZ794_28210) | 83703..84081 | + | 379 | Protein_102 | type-F conjugative transfer system protein TrbI | - |
| JZ794_RS28210 (JZ794_28215) | 84081..84707 | + | 627 | WP_004152590 | type-F conjugative transfer system protein TraW | traW |
| JZ794_RS28215 (JZ794_28220) | 84751..85710 | + | 960 | WP_015065634 | conjugal transfer pilus assembly protein TraU | traU |
| JZ794_RS28220 (JZ794_28225) | 85723..86361 | + | 639 | WP_004152682 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| JZ794_RS28225 (JZ794_28230) | 86409..88364 | + | 1956 | WP_040145764 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| JZ794_RS28230 (JZ794_28235) | 88396..88650 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| JZ794_RS28235 (JZ794_28240) | 88628..88876 | + | 249 | WP_013214029 | hypothetical protein | - |
| JZ794_RS28240 (JZ794_28245) | 88889..89215 | + | 327 | WP_013214030 | hypothetical protein | - |
| JZ794_RS28245 (JZ794_28250) | 89236..89988 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| JZ794_RS28250 (JZ794_28255) | 89999..90238 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| JZ794_RS28255 (JZ794_28260) | 90210..90767 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| JZ794_RS28260 (JZ794_28265) | 90813..91256 | + | 444 | WP_030003485 | F-type conjugal transfer protein TrbF | - |
| JZ794_RS28265 (JZ794_28270) | 91234..92613 | + | 1380 | WP_011977731 | conjugal transfer pilus assembly protein TraH | traH |
| JZ794_RS28270 (JZ794_28275) | 92613..95462 | + | 2850 | WP_004152624 | conjugal transfer mating-pair stabilization protein TraG | traG |
| JZ794_RS28275 (JZ794_28280) | 95475..96023 | + | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| JZ794_RS28280 (JZ794_28285) | 96207..96938 | + | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| JZ794_RS28285 (JZ794_28290) | 97130..97819 | + | 690 | WP_004198206 | hypothetical protein | - |
| JZ794_RS28290 (JZ794_28295) | 97948..100260 | + | 2313 | WP_043907039 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 5875 | GenBank | NZ_CP071282 |
| Plasmid name | pSA-KpST14-3 | Incompatibility group | IncFIB |
| Plasmid size | 166565 bp | Coordinate of oriT [Strand] | 71654..71703 [-] |
| Host baterium | Klebsiella pneumoniae subsp. pneumoniae strain SA-KpST14 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | silE, silS, silR, silC, silF, silB, silA, silP, pcoA, pcoB, pcoC, pcoD, pcoR, pcoE, arsH, arsC, arsB, arsA, arsD, arsR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |